Differential Gene Expression and Activation of Signaling Pathways in Radioresistant and Radiosensitive Cancer Cell Lines
The aim of the investigation was to study differential gene expression and signaling pathways activation in radioresistant and radiosensitive cancer cell lines.
Materials and Methods. K562 radioresistant cancer cell line of chronic myeloleukosis, and HCT-116p53 (+/+), HCT-116p53 (–/–), Me45 radiosensitive cancer cell lines were used in the study. Transcriptome analysis was performed using Affymetrix DNA microarray HGU133A series. Original Gene Selector program was used for bioinformation analysis. Protein interaction networks were studied by means of online STRING 9.0 system. To range the genes according to the signaling pathways the PANTHER program and QIAGEN SABiosciences database were used. Quantitative activation of signaling pathways was calculated with the help of OncoFinder bioinformation algorithm.
Results. Four genes (DAAM1, IFNAR2, PALLD, STK17A) were identified to increase their expression in radioresistant cell line and to decrease it in radiosensitive cell lines under radiation exposure. Three signal pathways — Wnt, Interferon and p53 — in which these genes are involved and which are common to the examined cell lines were found using PANTHER program. A significant difference in activation of these signaling pathways was discovered in radioresistant and radiosensitive cell lines.
Conclusion. Based on the analysis of molecular interaction networks and signaling pathway activation it has been revealed that the greatest differences between radioresistant and radiosensitive cell lines are observed in the activity of Interferon and p53 signaling pathways. TP53 gene expression level is not associated with the activity of p53 signaling pathway. Comparison of gene expression changes and activation of signaling pathways allows us to suggest the expression of DAAM1, IFNAR2, PALLD genes as indicators of radioresistance under radiation exposure.
Resistance of malignant tumors to radiation therapy remains the challenge that clinical and experimental oncology has to face. In addition to the originally inherent low sensitivity of some tumor cells to ionizing radiation, they are capable to acquire it in the course of therapy [1]. This can be explained by the fact, that cancer cells after radiation are able to endure DNA damage [2]. This results in their survival increase, causing infinite growth of cancer tumors and clinical progression of the disease. Besides, genetic instability of the tumor cells characterized by a high level of spontaneous mutations makes the situation more complicated. Therefore, further radiotherapy procedures do not improve the condition of oncological patients, and may lead to serious consequences, aggravating patient general condition and quality of life. This situation demands searching new paths to improve radiation therapy — overcoming radioresistance of cancer cells.
Radioresistance of cancer cells depends mainly on correct functioning of intracellular signaling pathways [3]. Alteration in one of the signaling pathway components, e.g. gene overexpression, is able to derange the functioning of the whole cell system. For example, ErbB-2 receptor, which is a member of the family of epidermal growth factor receptor (EGFR), is one of the receptors which are expressed most frequently on the cancer cell surface of bronchus epithelium. For EGFR gene, ErbB-2 acts as a coreceptor at the moment of forming dimer complex and later autophosphorylation, but the ligand has not been defined as yet [4]. Overexpression has been found to be characteristic of EGFR gene in malignant human neoplasms. As it is noted in various studies [5–7], there exists association between EGFR gene expression and resistance of cancer cells to radiotherapy. In the works [5–7] prostate cancer cells (DU145 and PC-3) are shown to express EGFR gene. Besides, interrelation of EGFR gene expression with proliferation, angiogenesis, migration of tumor cells and tumor radioresistance was found in preclinical investigations [8, 9]. Information on activation of signaling pathways and their components in response to radiotherapy will allow physicians to predict the outcomes of oncological disease treatment and to develop the method of suppressing radioresistance mechanism in cancer cells. Methods of full-transcriptome analysis give the most complete picture of the mechanisms of intracellular signaling pathways activation [10]. In the present work transcriptome analysis of radioresistant and radiosensitive cell lines was used to detect differentially expressing genes and investigate the differential activity of signaling pathways.
The aim of the investigation was to study differential gene expression and signaling pathways activation in radioresistant and radiosensitive cancer cell lines.
Materials and Methods
Cell cultures. The following cells were used in the experiment: K562 radioresistant suspension line of human erythroleukemic lymphoblastoid cells; HCT-116p53 (+/+) radiosensitive cell line of human rectal cancer with a normal TP53 gene; HCI-116p53 (–/–) radiosensitive cell line of human rectal cancer with a mutant TP53 gene; ME45 radiosensitive human melanoma cell line. Cell lines were obtained from American Type Culture Collection (ATCC).
Cells were cultured under standard conditions: in CO2-incubator (MCO-18AIC CO2, Helicon, Japan) at 37°C in 5% CO2 atmosphere and 98% humidity. RPMI-1640 medium containing L-glutamin with 10% fetal bovine serum and 5 µg/ml gentamicin was used for cultivation of K562 cell line. To cultivate HCT-116p53 (+/+), HCT-116p53 (–/–), Me45 cell lines DMEM/F12 medium also containing L-glutamin with 10% fetal bovine serum and 5 µg/ml gentamicin was used.
Experiment design. The cell lines were radiated in the logarithmic phase of growth. The cells were exposed a single time to X-rays generated by therapeutic accelerator (Clinac 600, Varian Medical Systems, USA) at a room temperature at a dose of 4 Gy [11, 12]. Irradiation dose rate amounted to 0.03 Gy/s at 104 cm focus distance. The height of water column over the cells was 1 cm. The cells were irradiated in 24-well plates (2.5 ml well volume). Gene expression analysis was performed 1, 12, and 24 h after the exposure.
Gene expression analysis. RNA was isolated from 3·106 cells using RNA isolation kit according to the manufacturer instructions. Integrity of the isolated RNA was tested by means of bioanalyzer (Agilent 2100, Agilent Technologies, USA). Cloned DNA library was prepared with the help of GeneChip Expression 3’-Amplification One-Cycle cDNA Synthesis Kit (Affymetrix, USA). Biotin labeling of antisense cloned RNA (cRNA) libraries and cleaning were performed using GeneChip Expression 3’-Amplification Reagents for IVT Labeling as indicated in manufacturer protocol. The quantity of RNA and DNA was estimated by a spectrometer (NanoDrop, Thermo Scientific, USA). Fragmentation of cRNA was conducted at 94°C in a thermocycler during 35 min. Synthetized biotinilated cRNA were first hybridized with Test-3 control matrix in order to assess the quality of the cRNA obtained. If the quality satisfied the rated one, hybridization with HGU133A was performed. The matrix was stained with streptavidin-phycoerythrin. The stained matrix was washed to eliminate the unconjugated protein and scanned on a scanner (GeneArray G2500A, Hewlett Packard, USA).
Bioinformatics analysis. Selection of genes whose expression changed relative to the control throughout the experiment, and search of common genes in radioresistant and radiosensitive cell lines was performed using original Gene Selector program (the program may be provided by the authors of the article), significance of differences was set at p≤0.05.
The network of molecular interaction was built by means of online STRING system (Search Tool for the Retrieval of Interacting Genes/Proteins). Data are presented based on four sources: Genomic Context, High-throughput Experiments, (Conserved) Coexpression, Previous Knowledge.
The signaling pathways, which the selected genes refer to, were determined using PANTHER program and QIAGEN SABiosciences database.
Quantitative activation of signaling pathways, which are influenced by the selected genes, was calculated with the help of OncoFinder method [13]. The degree of signaling pathways activation was measured in standard units.
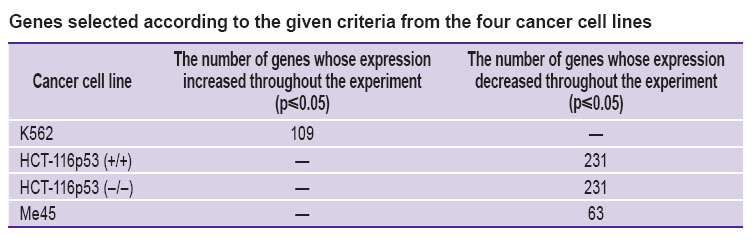
Results. As a result of statistical processing of the gene expression data of K562 cell line, 109 genes, whose expression increased throughout the experiment (See the Table), have been selected.
 Genes selected according to the given criteria from the four cancer cell lines Genes selected according to the given criteria from the four cancer cell lines
|
In the radiosensitive cancer cell lines with TP53 HCT-116p53 (+/+) normal gene and TP53 HCT-116p53 (–/–) mutant gene, 231 genes were isolated, while in Me45 cancer cell line, 63 genes whose expression decreased 1, 12, and 24 h after the experiment were isolated.
The Gene Selector program of automatic selection of genes with a given level of significance and expression selected four common genes for the cancer cell lines used: DAAM1, IFNAR2, PALLD, STK17A, whose expression increases in radioresistent cell line and decreases in radiosensitive one.
The selected genes were loaded in the online system STRING for detection of molecular interaction networks (Figure 1).
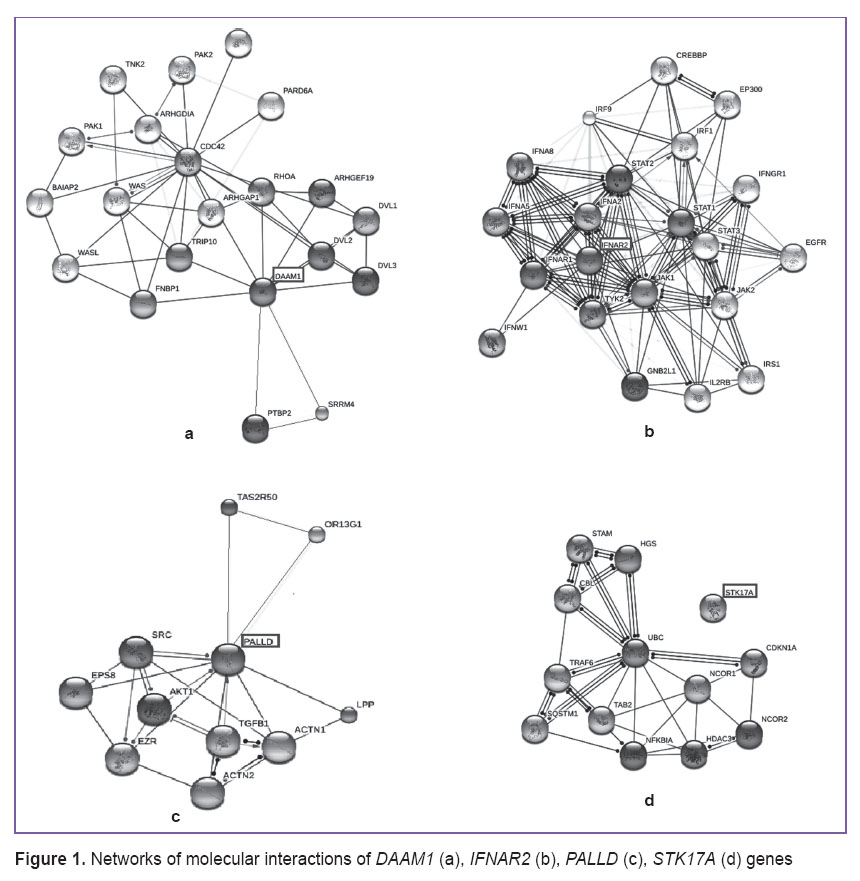 Figure 1. Networks of molecular interactions of DAAM1 (а), IFNAR2 (b), PALLD (c), STK17A (d) genes Figure 1. Networks of molecular interactions of DAAM1 (а), IFNAR2 (b), PALLD (c), STK17A (d) genes
|
DAAM1 gene has been estimated to interact directly with FNBP1, TRIP10, ARHGAP1, DVL1, DVL2, DVL3, PTBP2, RHOA, SRRM4 genes, through which the connection with ARHGDIA, BAIAP2, CDC42, PAK1, PAK2, PARD6A, PTBP2, TNK2, TRIP10, WAS, WASL genes is realized. IFNAR2 gene interacts with TYK2, GNB2L1, IFNA8, STAT1, IFNW1, STAT2, JAK1, IFNAR1, JAK2, STAT3, IFNA5 genes, through which it affects IL2RB, IRS1, EGFR, IFNGR1, IRF1, EP300, CREBBP, IRF9 genes. PALLD gene interacts with TGFB1, EPS8, AKT1, ACTN1, LPP, ACTN2, SRC, EZR genes.
When studying the influence of STK17A gene, there was detected a network of molecular interactions, where it does not affect other genes directly. This can be explained by the fact, that this gene is not sufficiently studied and the STRING program contains little information about its functional links with other genes.
Separate gene expression is liable to a wide range of variations and not always is an evidence of pathological process in contrast to the activity of the whole signaling pathway. Therefore, the selected DAAM1, IFNAR2, PALLD genes were downloaded to the PANTHER database, which contained information on the intracellular signaling pathways where they function (Figure 2).
 |
Figure 2. A number of signaling pathways which are influenced by DAAM1, IFNAR2, PALLD genes functioning in four cancer cell lines (K562, HCT-116p53 (+/+), HCT-116p53 (–/–), Me45) |
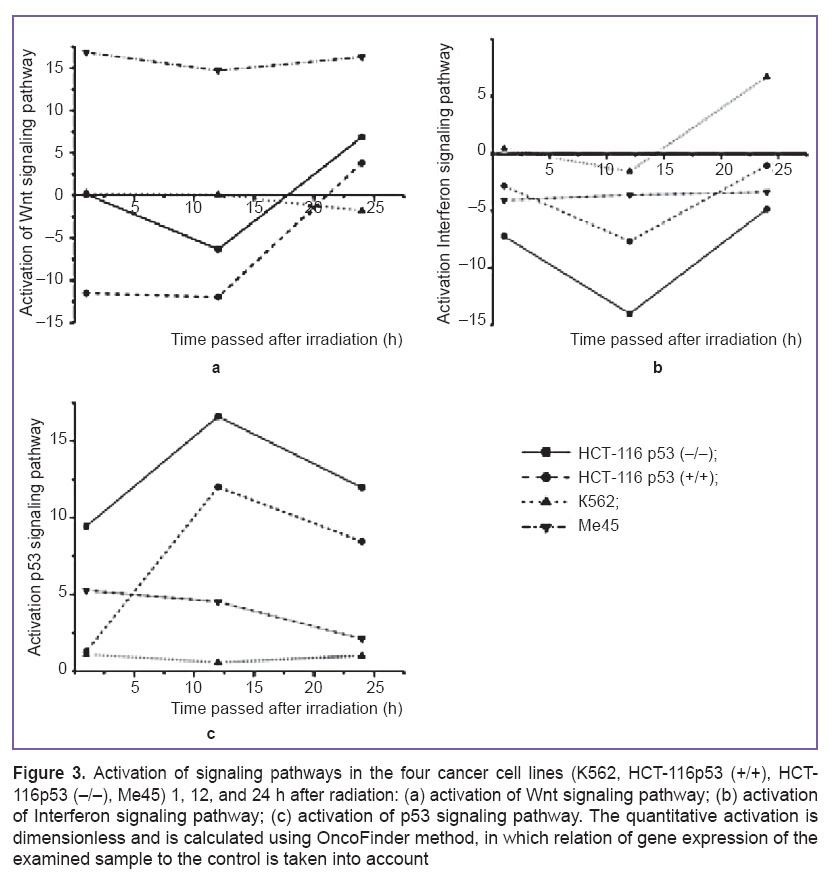
On the basis of the information obtained three signaling pathways were found (See Figure 2), which are common for radioresistance and radiosensitive cancer cell lines: Wnt signaling pathway, Interferon signaling pathway, p53 signaling pathway. By means of OncoFinder method their activation in the cell lines, which were directly affected by DAAM1, IFNAR2, PALLD, and STK17A genes, was calculated (Figure 3).
Activation in the HCT-116p53 (+/+), HCT-116p53 (–/–), and Me45 lines before 12 h decreases and then grows, as seen on the diagrams, reflecting the effect of ionizing radiation at a dose of 4 Gy on the activation of Wnt signaling pathway (Figure 3 (a)). In the HCT-116p53 (+/+) and HCT-116p53 (–/–) lines, values of activation after 24 h are significantly lower than in Me45 line. In radioresistant K562 cancer cell line activation of this signaling pathway decreases throughout the experiment.
The diagrams, reflecting the effect of ionizing radiation at a dose of 4 Gy on the activation ofInterferon signaling pathways (Figure 3 (b)), demonstrate, that despite a similar activation character in К562, HCT-116p53 (–/–), HCT-116p53 (+/+) cell lines, its values in these lines differ markedly.
Activation ofInterferon signaling pathway reaches its greatest value in the radioresistant cancer cell line after 24 h, while in radiosensitive lines its relatively low values are preserved till the end of the experiment.
As it is seen from the figure, activation of p53 signaling pathway in radioresistant and radiosensitive cell line is also different (Figure 3 (c)). In HCT-116p53 (+/+) and HCT-116p53 (–/–) it reaches its maximal value after 12 h from the beginning of the experiment, and it falls by 24 h. In K562 and Me45 cell lines activation equally decreases, but after 12 h it starts growing, while in Me45, on the contrary, it continues to fall.
Discussion. Intracellular signaling pathways, which include a great number of proteins, e.g. p53, phospholipase δ1, protein kinase C, clasterin and others, play an important role in generating the tumor cell response to ionizing radiation [14]. However, which particular signaling pathways participate in formation of low radiosensitivity of cancer cells remains a controversial question. Our investigations showed, that DAAM1, IFNAR2, PALLD, STK17A genes of radioresistant K562 cell line are characterized by overexpression in contrast to the genes of radiosensitive cell lines. DAAM1 gene is a member of formin protein family. It regulates cell growth and participates in generation and stretching of new actin filaments. It has been previously shown [15], that some actin-binding proteins, e.g. cofilin-1 (CFL1), are considerably activated in radioresistant astrocytoma.
IFNAR2 gene stimulates binding and activation of protein kinase Janus receptor, which phosphorylates several proteins (for example, STAT and STAT1). Signaling pathways of phosphatidylinositol-3-kinase (PI3K), protein kinase B (Act), Janus-kinase (JAK), transcription activator (STAT) are able to block apoptosis, which makes tumor cells resistant to radiotherapy [16, 17]. Rahaman et al. demonstrated in their studies, that inhibition of STAT3 signaling pathway results in intensive apoptosis and inhibition of proliferation in malignant glioma [18].
PALLD gene participates in organization of cell cytoskeleton, plays an important role in formation of morphology, motility and adhesion of the cells. Adhesion to extracellular matrix in normal and tumor cell lines promotes resistance to ionizing radiation [19–21].
STK17A gene is a constituent part of DAP-kinases and induces apoptosis. It regulates active oxygen forms in the cell. Increase of active oxygen forms in the cell can lead to adaptive changes and may play a central role in forming radioresistance in tumor cells [22].
Thus, DAAM1, IFNAR2, PALLD, and STK17A genes are involved in forming radioresistance and may be used as targets for the increase of radiosensitivity of tumor cells.
All mentioned genes in the radioresistant cell line are characterized by overexpression. Not only expression of separate genes should be taken into consideration in formation of resistance, but activation of signaling pathways, in which they participate. We have determined common signaling pathways for K562, HCT-116p53 (+/+), HCT-116p53 (–/–), Me45 cell lines, in which DAAM1, IFNAR2, PALLD, and STK17A genes are involved (See Figure 2).
In K562 radioresistant cell line activation of Wnt, Interferon, and p53 signaling pathways differed considerably from their activation in radiosensitive cell lines (See Figure 3 (b)).
It has been previously demonstrated [23], that Interferon signaling pathway is activated in radioresistant cancer cells. The key protein of this signaling pathway is a protein coded by STAT1 gene. IFNAR2 interacts physically with STAT1 protein. Zhan et al. showed, that overexpression of STAT1 promotes radioresistance of breast cancer cells [24]. Myeloma cells with STAT1 overexpression have been proved by Fryknäs et al. to be more radioresistant than the cells with a low expression level of this protein [25]. In our experiments increase of STAT1 gene expression was observed in all cell lines throughout the experiment, while overexpression of IFNAR2 gene was observed only in K562 radioresistant cell line during the experiment. Thus, IFNAR gene expression may serve as an indicator of radioresistance.
Many human malignant tumors contain mutations in p53 gene, and the majority of them have genetic damages in p53 signaling pathway [26]. For example, uveal melanoma has sometimes mutations in p53 gene, which prohibit normal functioning of p53 signaling pathway [27–29]. In this connection, uveal melanoma is capable to acquire low radiosensitivity. In our experiments we used two cell lines with a mutant TP53 gene: this is a radiosensitive HCT-116p53 (–/–) cell line and radioresistant К562 cell line. In both lines the protein p53 is not active. Nevertheless, activation of p53 signaling pathway in these cell lines is different. In K562 line radiation exposure does not affect the activity of this pathway throughout the experiment (See Figure 3 (c)). In radiosensitive HCT-116p53 (–/–) cell line activity of p53 signaling pathway in 1 h remains unchanged, but in 12 and 24 h it increases, though its level is somewhat lower than in the isogenic HCT-116p53 (+/+) cell line with TP53 wild-type gene. The data of our experiment demonstrate, that the value of TP53 gene expression is not associated with the activity of p53 signaling pathway.
Conclusion. For the first time, DAAM1, IFNAR2, PALLD, and STK17A genes are demonstrated to be associated with the phenomenon of cancer cell radioresistance. The greatest differences between radioresistant and radiosensitive cell lines are observed in the activity of Interferon and p53 signaling pathways. The value of TP53 gene expression is not associated with the activity of p53 signaling pathway.
Study Funding. The work was supported by the Ministry of Education and Science of Russian Federation (State task 6.1617.2014/K) and competition “Member of the Youth Scientific and Innovation Competition” No.2994GU1/2014.
Conflicts of Interest. The authors declare no conflicts of interest related to this study.
References
- Zinchenko V.A., Chaschina L.I. Possible mechanisms of the stability of tumor cells to radial and chemotherapy. Biopolym Cell 2005; 21(6): 473–484.
- Tang H.L., Tang H.M., Mak K.H., Hu S., Wang S.S., Wong K.M., Wong C.S., Wu H.Y., Law H.T., Liu K., Talbot C.C. Jr., Lau W.K., Montell D.J., Fung M.C. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell 2012; 23(12): 2240–2252, http://dx.doi.org/10.1091/mbc.E11-11-0926.
- Panganiban R.-A.M., Snow A.L., Day R.M. Mechanisms of radiation toxicity in transformed and non-transformed cells. Int J Mol Sci 2013; 14(8): 15931–15958, http://dx.doi.org/10.3390/ijms140815931.
- Tikhomirova A.V., Yagudina R.I. Pharmacoeconomic analysis of direct medical costs of metastatic colorectal cancer as the 1st line treatment regimes XELOX or FOLFOX and XELOX + BV or FOLFOX + BV. Farmakoekonomika 2010; 2: 22–27.
- Chinnaiyan P., Varambally S., Tomlins S.A., Ray S., Huang S., Chinnaiyan A.M., Harari P.M. Enhancing the antitumor activity of ErbB blockade with histone deacetylase (HDAC) inhibition. Int J Cancer 2006; 118(4): 1041–1050, http://dx.doi.org/10.1002/ijc.21465.
- Corvò R., Pastrone I., Scolaro T., Marcenaro M., Berretta L., Chiara S. Radiotherapy and oral capecitabine in the preoperative treatment of patients with rectal cancer: rationale, preliminary results and perspectives. Tumor 2003; 89(4): 361–367.
- El Sheikh S.S., Domin J., Abel P., Stamp G., Lalani El-N. Phosphorylation of both EGFR and ErbB2 is a reliable predictor of prostate cancer cell proliferation in response to EGF. Neoplasia 2004; 6(6): 846–853, http://dx.doi.org/10.1593/neo.04379.
- Milas L., Fan Z., Andratschke N.H., Ang K.K. Epidermal growth factor receptor and tumor response to radiation: in vivo preclinical studies. Int J Radiat Oncol Biol Phys 2004; 58(3): 966–971, http://dx.doi.org/10.1016/j.ijrobp.2003.08.035.
- Nasu S., Ang K.K., Fan Z., Milas L. C225 antiepidermal growth factor receptor antibody enhances tumor radiocurability. Int J Radiat Oncol Biol Phys 2001; 51(2): 474–477, http://dx.doi.org/10.1016/S0360-3016(01)01671-6.
- Lezhnina K., Kovalchuk O., Zhavoronkov A.A., Korzinkin M.B., Zabolotneva A.A., Shegay P.V., Sokov D.G., Gaifullin N.M., Rusakov I.G., Aliper A.M., Roumiantsev S.A., Alekseev B.Y., Borisov N.M., Buzdin A.A. Novel robust biomarkers for human bladder cancer based on activation of intracellular signaling pathways. Oncotarget 2014; 5(19): 9022–9032, http://dx.doi.org/10.18632/oncotarget.2493.
- Saenko Y.V., Shutov A.M., Rastorgueva E.V., Maslakova A.G. The role of mitochondria in the development of radiation-induced oxidative stress in K562 leukemia cells. Bull Exp Biol Med 2013; 154(5): 631–634, http://dx.doi.org/10.1007/s10517-013-2017-4.
- Spary L.K., Al-Taei S., Salimu J., Cook A.D., Ager A., Watson H.A., Clayton A., Staffurth J., Mason M.D., Tabi Z. Enhancement of T cell responses as a result of synergy between lower doses of radiation and T cell stimulation. J Immunol 2014; 192(7): 3101–3110, http://dx.doi.org/10.4049/jimmunol.1302736.
- Buzdin A.A., Zhavoronkov A.A., Korzinkin M.B., Venkova L.S., Zenin A.A., Smirnov P.Y., Borisov N.M. Oncofinder, a new method for the analysis of intracellular signaling pathway activation using transcriptomic data. Front Genet 2014; 5: 55, http://dx.doi.org/10.3389/fgene.2014.00055.
- Matsumoto H., Hamada N., Takahashi A., Kobayashi Y., Ohnishi T. Vanguards of paradigm shift in radiation biology: radiation-induced adaptive and bystander responses. J Radiat Res 2007; 48(2): 97–106, http://dx.doi.org/10.1269/jrr.06090.
- Yan H., Yang K., Xiao H., Zou Y.J., Zhang W.B., Liu H.Y. Over-expression of cofilin-1 and phosphoglycerate kinase 1 in astrocytomas involved in pathogenesis of radioresistance. CNS Neurosci Ther 2012; 18(9): 729–736, http://dx.doi.org/10.1111/j.1755-5949.2012.00353.x.
- Toker A., Yoeli-Lerner M. Akt signaling and cancer: surviving but not moving on. Cancer Res 2006; 66(8): 3963–3966, http://dx.doi.org/10.1158/0008-5472.CAN-06-0743.
- Hodge D.R., Hurt E.M., Farrar W.L. The role of IL-6 and STAT3 in inflammation and cancer. Eur J Cancer 2005;41(16): 2502–2512, http://dx.doi.org/10.1016/j.ejca.2005.08.016.
- Rahaman S.O., Harbor P.C., Chernova O., Barnett G.H., Vogelbaum M.A., Haque S.J. Inhibition of constitutively active STAT3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 2002; 21(55): 8404–8413, http://dx.doi.org/10.1038/sj.onc.1206047.
- Hehlgans S., Haase M., Cordes N. Signalling via integrins: implications for cell survival and anticancer strategies. Biochim Biophys Acta 2007; 1775(1): 163–180, http://dx.doi.org/10.1016/j.bbcan.2006.09.001.
- Eke I., Sandfort V., Storch K., Baumann M., Röper B., Cordes N. Pharmacological inhibition of EGFR tyrosine kinase affects ILK-mediated cellular radiosensitization in vitro. Int J Radiati Biol 2007; 83(11–12): 793–802, http://dx.doi.org/10.1080/09553000701727549.
- Park C.C., Zhang H., Pallavicini M., Gray J.W., Baehner F., Park C.J., Bissell M.J. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res 2006; 66(3): 1526–1535, http://dx.doi.org/10.1158/0008-5472.CAN-05-3071.
- Shi X., Zhang Y., Zheng J., Pan J. Reactive oxygen species in cancer stem cells. Antioxid Redox Signal 2012; 16(11): 1215–1228, http://dx.doi.org/10.1089/ars.2012.4529.
- Woodward W.A., Chen M.S., Behbod F., Alfaro M.P., Buchholz T.A., Rosen J.M. WNT/beta — catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad SciUSA2007; 104(2): 618–623, http://dx.doi.org/10.1073/pnas.0606599104.
- Zhan J.F., Chen L.H., Yuan Y.W., Xie G.Z., Sun A.M., Liu Y., Chen Z.X. STAT1 promotes radioresistance of CD44(+)/CD24(–/low) cells in breast cancer. Exp Biol Med (Maywood) 2011; 236(4): 418–22, http://dx.doi.org/10.1258/ebm.2011.010287.
- Fryknäs M., Dhar S., Oberg F., Rickardson L., Rydåker M., Göransson H., Gustafsson M., Pettersson U., Nygren P., Larsson R., Isaksson A. STAT1 signaling is associated with acquired crossresistance to doxorubicin and radiation in myeloma cell lines. Int J Cancer 2007; 120(1): 189–195, http://dx.doi.org/10.1002/ijc.22291.
- Sherr C.J. Principles of tumor suppression. Cell 2004; 116(2): 235–246, http://dx.doi.org/10.1016/S0092-8674(03)01075-4.
- Likhvantseva V.G. Prospects for developing treatment of uveal melanoma from the position of modern carcinogenesis concepts. Vestn Oftalmol 2002; 118(1): 32–35.
- Hussein M.R., Haemel A.K., Wood G.S. p53-related pathways and the molecular pathogenesis of melanoma. Eur J Cancer Prev 2003; 12(2): 93–100, http://dx.doi.org/10.1097/00008469-200304000-00002.
- Hussein M.R. The relationships between p53 protein expression and the clinicopathological features in the uveal melanomas. Cancer Biol Ther 2005; 4(1): 57–59, http://dx.doi.org/10.4161/cbt.4.1.1480.