Молекулярные механизмы аберрантной нейропластичности при заболеваниях аутистического спектра (обзор)
В обзоре проанализированы и систематизированы актуальные сведения о молекулярных механизмах развития расстройств аутистического спектра (РАС). Доказаны полиэтиологичность и многофакторность РАС. Осуществлена попытка совместного рассмотрения и систематизации современных гипотез патогенеза РАС на молекулярном уровне с позиции аберрантной пластичности головного мозга. Подробно рассмотрен механизм формирования глутаматной эксайтотоксичности, влияния на формирование РАС дисбаланса нейроактивных аминокислот и их производных, нейромедиаторов и гормонов. Проанализированы преимущества и недостатки предложенных гипотез с позиции доказательной медицины. Сделано заключение о ведущей роли глутаматной эксайтотоксичности как биохимического механизма аберрантной нейропластичности, сопровождающейся развитием окислительного стресса и митохондриальной дисфункцией. Прослежен механизм аберрантной нейропластичности в критические моменты развития нервной системы с учетом воздействия различных факторов внутренней и внешней среды. Рассмотрены новые подходы к поиску молекулярных маркеров РАС.
- Hrabovska S.V., Salyha Y.T. Animal models of autism spectrum disorders and behavioral techniques of their examination. Neurophysiology 2016; 48(5): 380–388, https://doi.org/10.1007/s11062-017-9613-2.
- Симашкова Н.В., Клюшник Т.П., Коваль-Зайцев А.А., Якупова Л.П. Клинико-биологические подходы к диагностике детского аутизма и детской шизофрении. Аутизм и нарушения развития 2016; 14(4): 51–67, https://doi.org/10.17759/autdd.2016140408.
- Yekoyan K.G., Grigoryan A., Fereshetyan K., Yeremyan D. Advances in understanding the pathophysiology of autism spectrum disorders. Behav Brain Res 2017; 331: 92–101, https://doi.org/10.1016/j.bbr.2017.04.038.
- Sharma S.R., Gonda X., Tarazi F.I. Autism spectrum disorder: classification, diagnosis and therapy. Pharmacol Ther 2018; 190: 91–104, https://doi.org/10.1016/j.pharmthera.2018.05.007.
- Wang K., Li N., Xu M., Huang M., Huang F. Glyoxalase 1 inhibitor alleviates autism-like phenotype in a prenatal valproic acid-induced mouse model. ACS Chem Neurosci 2020; 11(22): 3786–3792, https://doi.org/10.1021/acschemneuro.0c00482.
- Трифонова Е.А., Хлебодарова Т.М., Грунтенко Н.Е. Аутизм как проявление нарушения молекулярных механизмов регуляции развития и функций синапсов. Вавиловский журнал генетики и селекции 2016; 20: 959–967, https://doi.org/10.18699/vj16.217.
- Almeida L.E.F., Wang L., Khaibullina A., Quezado Z.M.N. Nicotinic cholinergic system alterations and nitrous oxide exposure in a mouse model: a hypothesis for the pathobiology of autism spectrum disorder. Psychopharmacology (Berl) 2017; 234(2): 317–318, https://doi.org/10.1007/s00213-016-4457-9.
- Bryn V., Aass H.C.D., Skjeldal O.H., Isaksen J., Saugstad O.D., Ormstad H. Cytokine profile in autism spectrum disorders in children. J Mol Neurosci 2017; 61(1): 1–7, https://doi.org/10.1007/s12031-016-0847-z.
- Liang S., Wang X., Zou M.Y., Wang H., Zhou X., Sun C.H., Xia W., Wu L.J., Fujisawa T.X., Tomoda A. Family-based association study of ZNF533, DOCK4 and IMMP2L gene polymorphisms linked to autism in a northeastern Chinese Han population. J Zhejiang Univ Sci B 2014; 15(3): 264–271, https://doi.org/10.1631/jzus.b1300133.
- Data & statistics on autism spectrum disorder. CDC — Centers for Disease Control and Prevention; 2018; URL: https://www.cdc.gov/ncbddd/autism/data.html.
- Durkin M.S., Elsabbagh M., Barbaro J., Gladstone M., Happe F., Hoekstra R.A., Lee L.C., Rattazzi A., Stapel-Wax J., Stone W.L., Tager-Flusberg H., Thurm A., Tomlinson M., Shih A. Autism screening and diagnosis in low resource settings: challenges and opportunities to enhance research and services worldwide. Autism Res 2015; 8(5): 473–476, https://doi.org/10.1002/aur.1575.
- Cohen S., Conduit R., Lockley S.W., Rajaratnam S.M.W., Cornish K.M. The relationship between sleep and behavior in autism spectrum disorder (ASD): a review. J Neurodev Disord 2014; 6(1): 44, https://doi.org/10.1186/1866-1955-6-44.
- Constantin L. The role of microRNAs in cerebellar development and autism spectrum disorder during embryogenesis. Mol Neurobiol 2017; 54: 6944–6959, https://doi.org/10.1007/s12035-016-0220-9.
- Dufault R., Lukiw W.J., Crider R., Schnoll R., Wallinga D., Deth R. A macroepigenetic approach to identify factors responsible for the autism epidemic in the United States. Clin Epigenetics 2012; 4(1): 6, https://doi.org/10.1186/1868-7083-4-6.
- Piras I.S., Haapanen L., Napolioni V., Sacco R., Van de Water J., Persico A.M. Anti-brain antibodies are associated with more severe cognitive and behavioral profiles in Italian children with autism spectrum disorder. Brain Behav Immun 2014; 38: 91–99, https://doi.org/10.1016/j.bbi.2013.12.020.
- Baker M.K., Godler D.E., Bui M., Hickerton C., Rogers C., Field M., Amor D.J., Bretherton L. Exploring autism symptoms in an Australian cohort of patients with Prader-Willi and Angelman syndromes. J Neurodev Disord 2018; 10(1): 24, https://doi.org/10.1186/s11689-018-9242-0.
- Buivydaite R., Newton C.R., Prasauskiene A. Scoping review: autism research in baltic states — what is known and what is still to be studied. J Autism Dev Disord 2017; 4: 294–306, https://doi.org/10.1007/s40489-017-0114-4.
- Ceylan A.C., Citli S., Erdem H.B., Sahin I., Acar Arslan E., Erdogan M. Importance and usage of chromosomal microarray analysis in diagnosing intellectual disability, global developmental delay, and autism; and discovering new loci for these disorders. Mol Cytogenet 2018; 11: 54, https://doi.org/10.1186/s13039-018-0402-4.
- Jung C.R., Lin Y.T., Hwang B.F. Air pollution and newly diagnostic autism spectrum disorders: a population-based cohort study in Taiwan. PLoS One 2013; 8(9): e75510, https://doi.org/10.1371/journal.pone.0075510.
- Hosseinpour M., Mashayekhi F., Bidabadi E., Salehi Z. Neuropilin-2 rs849563 gene variations and susceptibility to autism in Iranian population: a case-control study. Metab Brain Dis 2017; 32(5): 1471–1474, https://doi.org/10.1007/s11011-017-0024-2.
- Khaled E.M., Meguid N.A., Bjørklund G., Gouda A., Bahary M.H., Hashish A., Sallam N.M., Chirumbolo S., El-Bana M.A. Altered urinary porphyrins and mercury exposure as biomarkers for autism severity in Egyptian children with autism spectrum disorder. Metab Brain Dis 2016; 31(6): 1419–1426, https://doi.org/10.1007/s11011-016-9870-6.
- Переверзева Д.С., Горбачевская Н.Л. Нейробиологические маркеры ранних стадий расстройств аутистического спектра. Журнал высшей нервной деятельности им. И.П. Павлова 2016; 66(3): 289–301, https://doi.org/10.7868/s0044467716030102.
- Nayar K., Sealock J.M., Maltman N., Bush L., Cook E.H., Davis L.K., Losh M. Elevated polygenic burden for autism spectrum disorder is associated with the broad autism phenotype in mothers of individuals with autism spectrum disorder. Biol Psychiatry 2020, https://doi.org/10.1016/j.biopsych.2020.08.029.
- Diagnostic and statistical manual of mental disorders (DSM-5). Washington: American Psychiatric Association; 2013; 947 p.
- Симашкова Н.В. Расстройства аутистического спектра у детей. M: Авторская академия; 2013; 264 с.
- Alexiou A., Soursou G., Chatzichronis S., Gasparatos E., Kamal M.A., Yarla N.S., Perveen A., Barreto G.E., Ashraf G.M. Role of GTPases in the regulation of mitochondrial dynamics in Alzheimer’s disease and CNS-related disorders. Mol Neurobiol 2018; 56(6): 4530–4538, https://doi.org/10.1007/s12035-018-1397-x.
- Albajara Sáenz A., Van Schuerbeek P., Baijot S., Septier M., Deconinck N., Defresne P., Delvenne V., Passeri G., Raeymaekers H., Slama H., Victoor L., Willaye E., Peigneux P., Villemonteix T., Massat I. Disorder-specific brain volumetric abnormalities in attention-deficit/hyperactivity disorder relative to autism spectrum disorder. PLoS One 2020; 15(11): e0241856, https://doi.org/10.1371/journal.pone.0241856.
- Yoshimura S., Kobayashi K., Ueno T., Miyagi T., Oishi N., Murai T., Fujiwara H. Autistic traits are associated with the functional connectivity of between-but not within-attention systems in the general population. BMC Neurosci 2020; 21(1): 49, https://doi.org/10.1186/s12868-020-00603-2.
- Al-Ghamdi M., Al-Ayadhi L., El-Ansary A. Selected biomarkers as predictive tools in testing efficacy of melatonin and coenzyme Q on propionic acid-induced neurotoxicity in rodent model of autism. BMC Neurosci 2014; 15: 34, https://doi.org/10.1186/1471-2202-15-34.
- El-Ansary A., Bjørklund G., Khemakhem A.M., Al-Ayadhi L., Chirumbolo S., Ben Bacha A. Metabolism-associated markers and childhood autism rating scales (CARS) as a measure of autism severity. J Mol Neurosci 2018; 65(3): 265–276, https://doi.org/10.1007/s12031-018-1091-5.
- Roberts J.E., Bradshaw J., Will E., Hogan A.L., McQuillin S., Hills K. Emergence and rate of autism in fragile X syndrome across the first years of life. Dev Psychopathol 2020; 32(4): 1335–1352, https://doi.org/10.1017/s0954579420000942.
- Wolff J.J., Piven J. Predicting autism risk in infancy. J Am Acad Child Adolesc Psychiatry 2020, https://doi.org/10.1016/j.jaac.2020.07.910.
- McCray A.T., Trevvett P., Frost H.R. Modeling the autism spectrum disorder phenotype. Neuroinformatics 2014; 12(2): 291–305, https://doi.org/10.1007/s12021-013-9211-4.
- Keemink J.R., Jenner L., Prunty J.E., Wood N., Kelly D.J. Eye movements and behavioural responses to gaze-contingent expressive faces in typically developing infants and infant siblings. Autism Res 2020, https://doi.org/10.1002/aur.2432.
- Sandgreen H., Frederiksen L.H., Bilenberg N. Digital interventions for autism spectrum disorder: a meta-analysis. J Autism Dev Disord 2020, https://doi.org/10.1007/s10803-020-04778-9.
- Belger A., Carpenter K.L.H., Yucel G.H., Cleary K.M., Donkers F.C.L. The neural circuitry of autism. Neurotox Res 2011; 20(3): 201–214, https://doi.org/10.1007/s12640-010-9234-7.
- Kang D.W., Adams J.B., Gregory A.C., Borody T., Chittick L., Fasano A., Khoruts A., Geis E., Maldonado J., McDonough-Means S., Pollard E.L., Roux S., Sadowsky M.J., Lipson K.S., Sullivan M.B., Caporaso J.G., Krajmalnik-Brown R. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 2017; 5(1): 10, https://doi.org/10.1186/s40168-016-0225-7.
- Fujiwara T., Morisaki N., Honda Y., Sampei M., Tani Y. Chemicals, nutrition, and autism spectrum disorder: a mini-review. Front Neurosci 2016; 10: 174, https://doi.org/10.3389/fnins.2016.00174.
- Hu C.C., Sun Y.J., Liu C.X., Zhou B.R., Li C.Y., Xu Q., Xu X. NSDHL-containing duplication at Xq28 in a male patient with autism spectrum disorder: a case report. BMC Med Genet 2018; 19(1): 192, https://doi.org/10.1186/s12881-018-0705-7.
- Álvarez-Iglesias V., Mosquera-Miguel A., Cuscó I., Carracedo Á., Pérez-Jurado L.A., Salas A. Reassessing the role of mitochondrial DNA mutations in autism spectrum disorder. BMC Medical Genetics 2011; 12: 50, https://doi.org/10.1186/1471-2350-12-50.
- Duan W., Wang K., Duan Y., Chu X., Ma R., Hu P., Xiong B. Integrated transcriptome analyses revealed key target genes in mouse models of autism. Autism Res 2020; 13(3): 352–368, https://doi.org/10.1002/aur.2240.
- Wiebe S., Meng X.Q., Kim S.H., Zhang X., Lacaille J.C., Aguilar-Valles A., Sonenberg N. The eIF4E homolog 4EHP (eIF4E2) regulates hippocampal long-term depression and impacts social behavior. Mol Autism 2020; 11(1): 92, https://doi.org/10.1186/s13229-020-00394-7.
- Chapman N.H., Bernier R.A., Webb S.J., Munson J., Blue E.M., Chen D.H., Heigham E., Raskind W.H., Wijsman E.M. Replication of a rare risk haplotype on 1p36.33 for autism spectrum disorder. Hum Gen 2018; 137(10): 807–815, https://doi.org/10.1007/s00439-018-1939-3.
- Choi H., Song J., Park G., Kim J. Modeling of autism using organoid technology. Mol Neurobiol 2017; 54(10): 7789–7795, https://doi.org/10.1007/s12035-016-0274-8.
- Chiurazzi P., Kiani A.K., Miertus J., Paolacci S., Barati S., Manara E., Stuppia L., Gurrieri F., Bertelli M. Genetic analysis of intellectual disability and autism. Acta Biomed 2020; 91(13-S), https://doi.org/10.23750/abm.v91i13-s.10684.
- Mehta S.Q., Nurmi E.L. Genetic pathways to autism spectrum disorders. Neuropsychiatry 2013; 3(2): 193–207, https://doi.org/10.2217/npy.13.16.
- Bailey A., Le Couteur A., Gottesman I., Bolton P., Simonoff E., Yuzda E., Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med 1995; 25(1): 63–67.
- Gilman S.R., Iossifov I., Levy D., Ronemus M., Wigler M., Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron 2011; 70(5): 898–907, https://doi.org/10.1016/j.neuron.2011.05.021.
- Hiraide T., Nakashima M., Yamoto K., Fukuda T., Kato M., Ikeda H., Sugie Y., Aoto K., Kaname T., Nakabayashi K., Ogata T., Matsumoto N., Saitsu H. De novo variants in SETD1B are associated with intellectual disability, epilepsy and autism. Hum Genet 2018; 137(1): 95–104, https://doi.org/10.1007/s00439-017-1863-y.
- Goddard M.N., van Rijn S., Rombouts S.A.R.B., Swaab H. White matter microstructure in a genetically defined group at increased risk of autism symptoms, and a comparison with idiopathic autism: an exploratory study. Brain Imaging Behav 2016; 10(4): 1280–1288, https://doi.org/10.1007/s11682-015-9496-z.
- Kourtian S., Soueid J., Makhoul N.J., Guisso D.R., Chahrour M., Boustany R.M. Candidate genes for inherited autism susceptibility in the Lebanese population. Sci Rep 2017; 7: 45336, https://doi.org/10.1038/srep45336.
- Fakhoury M. Autistic spectrum disorders: a review of clinical features, theories and diagnosis. Int J Dev Neurosci 2015; 43: 70–77, https://doi.org/10.1016/j.ijdevneu.2015.04.003.
- Fakhoury M. Imaging genetics in autism spectrum disorders: linking genetics and brain imaging in the pursuit of the underlying neurobiological mechanisms. Prog Neuropsychopharmacol Biol Psychiatry 2018; 80(Pt B): 101–114, https://doi.org/10.1016/j.pnpbp.2017.02.026.
- Mahfouz A., Ziats M.N., Rennert O.M., Lelieveldt B.P.F., Reinders M.J.T. Shared pathways among autism candidate genes determined by co-expression network analysis of the developing human brain transcriptome. J Mol Neurosci 2015; 57(4): 580–594, https://doi.org/10.1007/s12031-015-0641-3.
- Shih P.Y., Hsieh B.Y., Tsai C.Y., Lo C.A., Chen B.E., Hsueh Y.P. Autism-linked mutations of CTTNBP2 reduce social interaction and impair dendritic spine formation via diverse mechanisms. Acta Neuropathol Commun 2020; 8(1): 185, https://doi.org/10.1186/s40478-020-01053-x.
- Connacher R.J., DiCicco-Bloom E., Millonig J.H. Using human induced neural precursor cells to define early neurodevelopmental defects in syndromic and idiopathic autism. Curr Pharmacol Rep 2018; 4: 422–435, https://doi.org/10.1007/s40495-018-0155-0.
- Masini E., Loi E., Vega-Benedetti A.F., Carta M., Doneddu G., Fadda R., Zavattari P. An overview of the main genetic, epigenetic and environmental factors involved in autism spectrum disorder focusing on synaptic activity. Int J Mol Sci 2020; 21(21): 8290, https://doi.org/10.3390/ijms21218290.
- Farahani M., Rezaei-Tavirani M., Zali A., Zamanian-Azodi M. Systematic analysis of protein-protein and gene-environment interactions to decipher the cognitive mechanisms of autism spectrum disorder. Cell Mol Neurobiol 2020, https://doi.org/10.1007/s10571-020-00998-w.
- Hannon E., Schendel D., Ladd-Acosta C., Grove J.; iPSYCH-Broad ASD Group, Hansen C.S., Andrews S.V., Hougaard D.M., Bresnahan M., Mors O., Hollegaard M.V., Bækvad-Hansen M., Hornig M., Mortensen P.B., Børglum A.D., Werge T., Pedersen M.G., Nordentoft M., Buxbaum J., Daniele Fallin M., Bybjerg-Grauholm J., Reichenberg A., Mill J. Elevated polygenic burden for autism is associated with differential DNA methylation at birth. Genome Med 2018; 10(1): 19, https://doi.org/10.1186/s13073-018-0527-4.
- Gamsiz E.D., Sciarra L.N., Maguire A.M., Pescosolido M.F., Van Dyck L.I., Morrow R.M. Discovery of rare mutations in autism: elucidating neurodevelopmental mechanisms. Neurotherapeutics 2015; 12(3): 553–571, https://doi.org/10.1007/s13311-015-0363-9.
- Siddiqui M.F., Elwell C., Johnson M.H. Mitochondrial dysfunction in autism spectrum disorders. Autism Open Access 2016; 6(5): 1000190, https://doi.org/10.4172/2165-7890.1000190.
- Surén P., Gunnes N., Roth C., Bresnahan M., Hornig M., Hirtz D., Lie K.K., Lipkin W.I., Magnus P., Reichborn-Kjennerud T., Schjølberg S., Susser E., Øyen A.S., Smith G.D., Stoltenberg C. Parental obesity and risk of autism spectrum disorder. Pediatrics 2014; 133(5): e1128–e1138, https://doi.org/10.1542/peds.2013-3664.
- Veroniki A.A., Cogo E., Rios P., Straus S.E., Finkelstein Y., Kealey R., Reynen E., Soobiah C., Thavorn K., Hutton B., Hemmelgarn B.R., Yazdi F., D’Souza J., MacDonald H., Tricco A.C. Comparative safety of anti-epileptic drugs during pregnancy: a systematic review and network meta-analysis of congenital malformations and prenatal outcomes. BMC Med 2017; 15(1): 95, https://doi.org/10.1136/bmjopen-2017-017248.
- Kobayashi T., Matsuyama T., Takeuchi M., Ito S. Autism spectrum disorder and prenatal exposure to selective serotonin reuptake inhibitors: a systematic review and meta-analysis. Reprod Toxicol 2016; 65: 170–178, https://doi.org/10.1016/j.reprotox.2016.07.016.
- Wan H., Zhang C., Li H., Luan S., Liu C. Association of maternal diabetes with autism spectrum disorders in offspring. Medicine (Baltimore) 2018; 97(2): e9438, https://doi.org/10.1097/md.0000000000009438.
- Li M., Fallin M.D., Riley A., Landa R., Walker S.O., Silverstein M., Caruso D., Pearson C., Kiang S., Dahm J.L., Hong X., Wang G., Wang M.C., Zuckerman B., Wang X. The association of maternal obesity and diabetes with autism and other developmental disabilities. Pediatrics 2016; 137(2): e20152206, https://doi.org/10.1542/peds.2015-2206.
- Barkoski J.M., Philippat C., Tancredi D., Schmidt R.J., Ozonoff S., Barr D.B., Elms W., Bennett D., Hertz-Picciotto I. In utero pyrethroid pesticide exposure in relation to autism spectrum disorder (ASD) and other neurodevelopmental outcomes at 3 years in the MARBLES longitudinal cohort. Environ Res 2020; 194: 110495, https://doi.org/10.1016/j.envres.2020.110495.
- Yasuda H., Tsutsui T. Assessment of infantile mineral imbalances in autism spectrum disorders (ASDs). Int J Environ Res Public Health 2013; 10(11): 6027–6043, https://doi.org/10.3390/ijerph10116027.
- Skalny A.V., Simashkova N.V., Klyushnik T.P., Grabeklis A.R., Bjørklund G., Skalnaya M.G., Nikonorov A.A., Tinkov A.A. Hair toxic and essential trace elements in children with autism spectrum disorder. Metab Brain Dis 2017; 32(1): 195–202, https://doi.org/10.1007/s11011-016-9899-6.
- Skalny A.V., Skalnaya M.G., Bjørklund G., Nikonorov A.A., Tinkov A.A. Mercury as a possible link between maternal obesity and autism spectrum disorder. Med Hypotheses 2016; 91: 90–94, https://doi.org/10.1016/j.mehy.2016.04.021.
- Saldanha Tschinkel P.F., Bjørklund G., Conón L.Z.Z., Chirumbolo S., Nascimento V.A. Plasma concentrations of the trace elements copper, zinc and selenium in Brazilian children with autism spectrum disorder. Biomed Pharmacother 2018; 106: 605–609, https://doi.org/10.1016/j.biopha.2018.06.174.
- Jafari T., Rostampour N., Fallah A.A., Hesami A. The association between mercury levels and autism spectrum disorders: a systematic reviewed meta-analysis. J Trace Elem Med Biol 2017; 44: 289–297, https://doi.org/10.1016/j.jtemb.2017.09.002.
- Mostafa G.A., Bjørklund G., Urbina M.A., Al-Ayadhi L.Y. The levels of blood mercury and inflammatory-related neuropeptides in the serum are correlated in children with autism spectrum disorder. Metab Brain Dis 2016; 31(3): 593–599, https://doi.org/10.1007/s11011-015-9784-8.
- Saghazadeh A., Ahangari N., Hendi K., Saleh F., Rezaei N. Status of essential elements in autism spectrum disorder: systematic review and meta-analysis. Rev Neurosci 2017; 28(7): 783–809, https://doi.org/10.1515/revneuro-2017-0015.
- Hawari I., Eskandar M.B., Alzeer S. The role of lead, marganese, and zinc in autism spectrum disorders (ASDs) and attention-deficient hyperactivity disorder (ADHD): a case-control study on Syrian children affected by the Syrian crisis. Biol Trace Elem Res 2020; 197(1): 107–114, https://doi.org/10.1007/s12011-020-02146-3.
- Vyas Y., Lee K., Jung Y., Montgomery J.M. Influence of maternal zinc supplementation on the development of autism-associated behavioural and synaptic deficits in offspring Shank3-knockout mice. Mol Brain 2020; 13(1): 110, https://doi.org/10.1186/s13041-020-00650-0.
- Shih P.Y., Hsieh B.Y., Lin M.H., Huang T.N., Tsai C.Y., Pong W.L., Lee S.P., Hsueh Y.P. CTTNBP2 controls synaptic expression of zinc-related autism-associated proteins and regulates synapse formation and autism-like behaviors. Cell Rep 2020; 31(9): 107700, https://doi.org/10.1016/j.celrep.2020.107700.
- Colle D., Farina M., Ceccatelli S., Raciti M. Paraquat and maneb exposure alters rat neural stem cell proliferation by inducing oxidative stress: new insights on pesticide-induced neurodevelopmental toxicity. Neurotox Res 2018; 34(4): 820–833, https://doi.org/10.1007/s12640-018-9916-0.
- De Felice A., Greco A., Calamandrei G., Minghetti L. Prenatal exposure to the organophosphate insecticide chlorpyrifos enhances brain oxidative stress and prostaglandin E2 synthesis in a mouse model of idiopathic autism. J Neuroinflammation 2016; 13(1): 149, https://doi.org/10.1186/s12974-016-0617-4.
- Henriksen A.D., Andrade A., Harris E.P., Rissman E.F., Wolstenholme J.T. Bisphenol a exposure in utero disrupts hypothalamic gene expression particularly genes suspected in autism spectrum disorders and neuron and hormone signaling. Int J Mol Sci 2020; 21(9): 3129, https://doi.org/10.3390/ijms21093129.
- Zerbo O., Qian Y., Yoshida C., Fireman B.H., Klein N.P., Croen L.A. Association between influenza infection and vaccination during pregnancy and risk of autism spectrum disorder. JAMA Pediatr 2017; 171(1): e163609, https://doi.org/10.1001/jamapediatrics.2016.3609.
- Whiteley P., Shattock P., Knivsberg A.M., Seim A., Reichelt K.L., Todd L., Carr K., Hooper M. Gluten- and casein-free dietary intervention for autism spectrum conditions. Front Hum Neurosci 2013; 6: 344, https://doi.org/10.3389/fnhum.2012.00344.
- Чернов А.Н. Патофизиологические механизмы развития аутизма у детей. Журнал неврологии и психиатрии им. С.С. Корсакова 2020; 120(3): 97–108, https://doi.org/10.17116/jnevro202012003197.
- Hegarty J.P. II, Weber D.J., Cirstea C.M., Beversdorf D.Q. Cerebro-cerebellar functional connectivity is associated with cerebellar excitation–inhibition balance in autism spectrum disorder. J Autism Dev Disord 2018; 48(10): 3460–3473, https://doi.org/10.1007/s10803-018-3613-y.
- White S.W., Mazefsky C.A., Dichter G.S., Chiu P.H., Richey J.A., Ollendick T.H. Social-cognitive, physiological, and neural mechanisms underlying emotion regulation impairments: understanding anxiety in autism spectrum disorder. Int J Dev Neurosci 2014; 39: 22–36, https://doi.org/10.1016/j.ijdevneu.2014.05.012.
- Schaefer G.B., Medelsohn N.J.; Professional Practice and Guidelines Committee. Clinical genetic evaluation in identifying the etiology of autism spectrum disorders: guideline revisions. Genet Med 2013; 15(5): 399–407, https://doi.org/10.1038/gim.2013.32.
- Christensen J., Grønborg T.K., Sørensen M.J., Schendel D., Parner E.T., Pedersen L.H., Vestergaard M. Prenatal valproate exposure and risk of autism spectrum disorders and childhood autism. JAMA 2013; 309(16): 1696–1703, https://doi.org/10.1001/jama.2013.2270.
- Surén P., Roth C., Bresnahan M., Haugen M., Hornig M., Hirtz D., Lie K.K., Lipkin W.I., Magnus P., Reichborn-Kjennerud T., Schjølberg S., Davey Smith G., Øyen A.S., Susser E., Stoltenberg C. Association between maternal use of folic acid supplements and risk of autism spectrum disorders in children. JAMA 2013; 309(6): 570–577, https://doi.org/10.1001/jama.2012.155925.
- Sung M., Chin C.H., Lim C.G., Liew H.S.A., Lim C.S., Kashala E., Weng S.J. What´s in the pipeline? Drugs in development for autism spectrum disorder. Neuropsychiatr Dis Treat 2014; 10: 371–381, https://doi.org/10.2147/ndt.s39516.
- Ann Abraham D., Narasimhan U., Christy S., Muhasaparur Ganesan R. Effect of L-carnosine as adjunctive therapy in the management of children with autism spectrum disorder: a randomized controlled study. Amino Acids 2020; 52(11–12): 1521–1528, https://doi.org/10.1007/s00726-020-02909-1.
- Alfawaz H., Al-Onazi M., Bukhari S.I., Binobead M., Othman N., Algahtani N., Bhat R.S., Moubayed N.M.S., Alzeer H.S., El-Ansary A. The independent and combined effects of omega-3 and vitamin B12 in ameliorating propionic acid induced biochemical features in juvenile rats as rodent model of autism. J Mol Neurosci 2018; 66(3): 403–413, https://doi.org/10.1007/s12031-018-1186-z.
- Marotta R., Risoleo M.C., Messina G., Parisi L., Carotenuto M., Vetri L., Roccella M. The neurochemistry of autism. Brain Sci 2020; 10(3): 163, https://doi.org/10.3390/brainsci10030163.
- Essa M.M., Braidy N., Vijayan K.R., Subash S., Guillemin G.J. Excitotoxicity in the pathogenesis of autism. Neurotox Res 2013; 23(4): 393–400, https://doi.org/10.1007/s12640-012-9354-3.
- Ghanizadeh A. Targeting of glycine site on NMDA receptor as a possible new strategy for autism treatment. Neurochem Res 2011; 36(5): 922–923, https://doi.org/10.1007/s11064-010-0381-2.
- Al-Otaish H., Al-Ayadhi L., Bjørklund G., Chirumbolo S., Urbina M.A., El-Ansary A. Relationship between absolute and relative ratios of glutamate, glutamine and GABA and severity of autism spectrum disorder. Metab Brain Dis 2018; 33(3): 843–854, https://doi.org/10.1007/s11011-018-0186-6.
- Auger M.L., Meccia J., Galea L.A.M., Floresco S.B. Disinhibition of the prefrontal cortex leads to brain-wide increases in neuronal activation that are modified by spatial learning. Brain Struct Funct 2018; 224(1): 171–190, https://doi.org/10.1007/s00429-018-1769-z.
- Levin A.R., Nelson C.A. Inhibition-based biomarkers for autism spectrum disorder. Neurotherapeutics 2015; 12(3): 546–552, https://doi.org/10.1007/s13311-015-0350-1.
- Alabdali A., Al-Ayadhi L., El-Ansary A. Association of social and cognitive impairment and biomarkers in autism spectrum disorders. J Neuroinflammation 2014; 11: 4, https://doi.org/10.1186/1742-2094-11-4.
- Muller C.L., Anacker A.M.J., Veenstra-VanderWeele J. The serotonin system in autism spectrum disorder: from biomarker to animal models. Neuroscience 2016; 321: 24–41, https://doi.org/10.1016/j.neuroscience.2015.11.010.
- Amodeo D.A., Oliver B., Pahua A., Hitchcock K., Bykowski A., Tice D., Musleh A., Ryan B.C. Serotonin 6 receptor blockade reduces repetitive behavior in the BTBR mouse model of autism spectrum disorder. Pharmacol Biochem Behav 2021; 200: 173076, https://doi.org/10.1016/j.pbb.2020.173076.
- Israelyan N., Margolis K.G. Serotonin as a link between the gut-brain-microbiome axis in autism spectrum disorders. Pharmacol Res 2018; 132: 1–6, https://doi.org/10.1016/j.phrs.2018.03.020.
- Горина А.С. Биохимические механизмы развития аутизма и синдрома дефицита внимания и гиперактивности у детей. Автореф. дисс. … докт. биол. наук. Новосибирск; 2013.
- El-Ansary A.K., Ben Bacha A., Al-Ayahdi L.Y. Relationship between chronic lead toxicity and plasma neurotransmitters in autistic patients from Saudi Arabia. Clin Biochem 2011; 44(13): 1116–1120, https://doi.org/10.1016/j.clinbiochem.2011.06.982.
- Gevi F., Belardo A., Zolla L. A metabolomics approach to investigate urine levels of neurotransmitters and related metabolites in autistic children. Biochim Biophys Acta Mol Basis Dis 2020; 1866(10): 165859, https://doi.org/10.1016/j.bbadis.2020.165859.
- Строганова Т.А., Орехова Е.В., Галюта И.А. Нейронные механизмы нарушений ориентировки внимания у детей с расстройством аутистического спектра. Экспериментальная психология 2015; 8(3): 7–23, https://doi.org/10.17759/exppsy.2015080302.
- Wang X., Qiao Y., Dai Z., Sui N., Shen F., Zhang J., Liang J. Medium spiny neurons of the anterior dorsomedial striatum mediate reversal learning in a cell-type-dependent manner. Brain Struct Funct 2019; 224(1): 419–434, https://doi.org/10.1007/s00429-018-1780-4.
- Tolomeo S., Chiao B., Lei Z., Chew S.H., Ebstein R.P. A novel role of CD38 and oxytocin as tandem molecular moderators of human social behavior. Neurosci Biobehav Rev 2020; 115: 251–272, https://doi.org/10.1016/j.neubiorev.2020.04.013.
- Bjørklund G., Meguid N.A., El-Ansary A., El-Bana M.A., Dadar M., Aaseth J., Hemimi M., Osredkar J., Chirumbolo S. Diagnostic and severity-tracking biomarkers for autism spectrum disorder. J Mol Neurosci 2018; 66(4): 492–511, https://doi.org/10.1007/s12031-018-1192-1.
- Kohls G., Chevallier C., Troiani V., Schultz R.T. Social ‘wanting’ dysfunction in autism: neurobiological underpinnings and treatment implications. J Neurodev Disord 2012; 4(1): 10, https://doi.org/10.1186/1866-1955-4-10.
- Carson D.S., Garner J.P., Hyde S.A., Libove R.A., Berquist S.W., Hornbeak K.B., Jackson L.P., Sumiyoshi R.D., Howerton C.L., Hannah S.L., Partap S., Phillips J.M., Hardan A.Y., Parker K.J. Arginine vasopressin is a blood-based biomarker of social functioning in children with autism. PloS One 2015; 10(7): e0132224, https://doi.org/10.1371/journal.pone.0132224.
- Rutigliano G., Rocchetti M., Paloyelis Y., Gilleen J., Sardella A., Cappucciati M., Palombini M., Dell’Osso L., Caverzasi E., Politi P., McGuire P., Fusar-Poli P. Peripheral oxytocin and vasopressin: biomarkers of psychiatric disorders? A comprehensive systematic review and preliminary meta-analysis. Psychiatry Res 2016; 241: 207–220, https://doi.org/10.1016/j.psychres.2016.04.117.
- Chauhan F., Audhya T., Chauhan V. Brain region-specific glutathione redox imbalance in autism. Neurochem Res 2012; 37(8): 1681–1689, https://doi.org/10.1007/s11064-012-0775-4.
- Yui K., Kawasaki Y., Yamada H., Ogawa S. Oxidative stress and nitric oxide in autism spectrum disorder and other neuropsychiatric disorders. CNS Neurol Disord Drug Targets 2016; 15(5): 587–596, https://doi.org/10.2174/1871527315666160413121751.
- Gu F., Chauhan V., Kaur K., Brown W.T., LaFauci G., Wegiel J., Chauhan A. Alterations in mitochondrial DNA copy number and the activities of electron transport chain complexes and pyruvate dehydrogenase in the frontal cortex from subjects with autism. Transl Psychiatry 2013; 3(9): e299, https://doi.org/10.1038/tp.2013.68.
- Guevara-Campos J., Gonz á lez-Guevara L., Puig-Alcaraz C., Cauli O. Autism spectrum disorders associated to a deficiency of the enzymes of the mitochondrial respiratory chain. Metab Brain Dis 2013; 28(4): 605–612, https://doi.org/10.1007/s11011-013-9419-x.
- Полякова С.И. Патофизиологическое обоснование персонифицированной метаболической терапии РАС. Перспективные методы лечения. Аутизм и нарушения развития 2019; 17(1): 55–70, https://doi.org/10.17759/autdd.2019170106.
- Frackowiak J., Mazur-Kolecka B., Schanen N.C., Brown W.T., Wegiel J. The link between intraneuronal N-truncated amyloid-β peptide and oxidatively modified lipids in idiopathic autism and dup(15q11.2-q13)/autism. Acta Neuropathol Commun 2013; 1: 61, https://doi.org/10.1186/2051-5960-1-61.
- Ghanizadeh A. Malondialdehyde, Bcl-2, superoxide dismutase and glutathione peroxidase may mediate the association of sonic hedgehog protein and oxidative stress in autism. Neurochem Res 2012; 37(4): 899–901, https://doi.org/10.1007/s11064-011-0667-z.
- Manivasagam T., Arunadevi S., Essa M.M., SaravanaBabu C., Borah A., Thenmozhi A.J., Qoronfleh M.W. Role of oxidative stress and antioxidants in autism. Adv Neurobiol 2020; 24: 193–206, https://doi.org/10.1007/978-3-030-30402-7_7.
- Matsuo K., Yabuki Y., Fukunaga K. 5-aminolevulinic acid inhibits oxidative stress and ameliorates autistic-like behaviors in prenatal valproic acid-exposed rats. Neuropharmacology 2020; 168: 107975, https://doi.org/10.1016/j.neuropharm.2020.107975.
- Gillberg C., Fernell E., Kočovská E., Minnis H., Bourgeron N., Thompson L., Allely C.S. The role of cholesterol metabolism and various steroid abnormalities in autism spectrum disorders: a hypothesis paper. Autism Res 2017; 10(6): 1022–1044, https://doi.org/10.1002/aur.1777.
- Mostafa G.A., Al-Ayadhi L.Y. Reduced serum concentrations of 25-hydroxy vitamin D in children with autism: relation to autoimmunity. J Neuroinflammation 2012; 9: 201, https://doi.org/10.1186/1742-2094-9-201.
- Magnusson C., Kosidou K., Dalman C., Lundberg M., Lee B.K., Rai D., Karlsson H., Gardner R., Arver S. Maternal vitamin D deficiency and the risk of autism spectrum disorders: population-based study. BJ Psych Open 2016; 2: 170–172, https://doi.org/10.1192/bjpo.bp.116.002675.
- Pangrazzi L., Balasco L., Bozzi Y. Oxidative stress and immune system dysfunction in autism spectrum disorders. Int J Mol Sci 2020; 21(9): 3293, https://doi.org/10.3390/ijms21093293.
- Bjørklund G., Saad K., Chirumbolo S., Kern J.K., Geier D.A., Geier M.R., Urbina M.A. Immune dysfunction and neuroinflammation in autism spectrum disorder. Acta Neurobiol Exp (Wars) 2016; 76(4): 256–268, https://doi.org/10.21307/ane-2017-025.
- Mostafa G.A., Al-Ayadhi L.Y. The possible link between the elevated serum levels of neurokinin A and anti-ribosomal P protein antibodies in children with autism. J Neuroinflammation 2011; 8: 180, https://doi.org/10.1186/1742-2094-8-180.
- Ahmad S.F., Bakheet S.A., Ansari M.A., Nadeem A., Alobaidi A.F., Attia S.M., Alhamed A.S., Aldossari A.A., Mahmoud M.A. Methylmercury chloride exposure aggravates proinflammatory mediators and Notch-1 signaling in CD14+ and CD40+ cells and is associated with imbalance of neuroimmune function in BTBR T+ Itpr3tf/J mice. Neurotoxicology 2020; 82: 9–17, https://doi.org/10.1016/j.neuro.2020.10.014.
- Al-Ayadhi L.Y., Mostafa G.A. Serum antinucleosome-specific antibody as a marker of autoimmunity in children with autism. J Neuroinflammation 2014; 11: 69, https://doi.org/10.1186/1742-2094-11-69.
- Kern J.K., Geier D.A., Sykes L.K., Geier M.R. Evidence of neurodegeneration in autism spectrum disorder. Transl Neurodegener 2013; 2(1): 17, https://doi.org/10.1186/2047-9158-2-17.
- Ghaffari M.A., Mousavinejad E., Riahi F., Mousavinejad M., Afsharmanesh M.R. Increased serum levels of tumor necrosis factor-alpha, resistin, and visfatin in the children with autism spectrum disorders: a case-control study. Neurol Res Int 2016; 2016: 9060751, https://doi.org/10.1155/2016/9060751.
- Ahmad S.F., Zoheir K.M.A., Ansari M.A., Nadeem A., Bakheet S.A., Al-Ayadhi L.Y., Alzahrani M.Z., Al-Shabanah O.A., Al-Harbi M.M., Attia S.M. Dysregulation of Th1, Th2, Th17, and T regulatory cell-related transcription factor signaling in children with autism. Mol Neurobiol 2017; 54(6): 4390–4400, https://doi.org/10.1007/s12035-016-9977-0.
- Al-Ayadhi L.Y., Mostafa G.A. Elevated serum levels of interleukin-17A in children with autism. J Neuroinflammation 2012; 9: 158, https://doi.org/10.1186/1742-2094-9-158.
- Brigida A.L., Schultz S., Cascone M., Antonucci N., Siniscalco D. Endocannabinod signal dysregulation in autism spectrum disorders: a correlation link between inflammatory state and neuro-immune alterations. Int J Mol Sci 2017; 18(7): 1425, https://doi.org/10.3390/ijms18071425.
- Chew L., Sun K.L., Sun W., Wang Z., Rajadas J., Flores R.E., Arnold E., Jo B., Fung L.K. Association of serum allopregnanolone with restricted and repetitive behaviors in adult males with autism. Psychoneuroendocrinology 2020; 123: 105039, https://doi.org/10.1016/j.psyneuen.2020.105039.
- O’Connor R., van De Wouw M., Moloney G.M., Ventura-Silva A.P., O’Riordan K., Golubeva A., Dinan T.G., Schellekens H., Cryan J.F. Strain differences in behaviour and immunity in aged mice: relevance to autism. Behav Brain Res 2020; 399: 113020, https://doi.org/10.1016/j.bbr.2020.113020.
- Hsiao E.Y. Gastrointestinal issues in autism spectrum disorder. Harv Rev Psychiatry 2014; 22(2): 104–111, https://doi.org/10.1097/hrp.0000000000000029.
- Samsam M., Ahangari R., Naser S.A. Pathophysiology of autism spectrum disorders: revisiting gastrointestinal involvement and immune imbalance. World J Gastroenterol 2014; 20(29): 9942–9951, https://doi.org/10.3748/wjg.v20.i29.9942.
- Бавыкина И.А., Попов В.И., Звягин А.A. Бавыкин Д.В. Частота выявления маркеров непереносимости казеина и глютена у детей с расстройствами аутистического спектра. Bопросы питания 2019; 88(4): 41–47, https://doi.org/10.24411/0042-8833-2019-10040.
- Xiong C., Sun S., Jiang W., Ma L., Zhang J. ASDmiR: a stepwise method to uncover miRNA regulation related to autism spectrum disorder. Front Genet 2020; 11: 562971, https://doi.org/10.3389/fgene.2020.562971.
- Enriquez-Barreto L., Morales M. The PI3K signaling pathway as a pharmacological target in autism related disorders and schizophrenia. Mol Cell Ther 2016; 4: 2, https://doi.org/10.1186/s40591-016-0047-9.
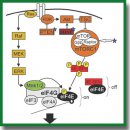
- Amorim I.S., Lach G., Gkogkas C.G. The role of the eukaryotic translation initiation factor 4E (eIF4E) in neuropsychiatric disorders. Front Genet 2018; 9: 561, https://doi.org/10.3389/fgene.2018.00561.
- Lv J.W., Cheng T.L., Qiu Z.L., Zhou W.H. Role of the PTEN signaling pathway in autism spectrum disorder. Neurosci Bull 2013; 29(6): 773–778, https://doi.org/10.1007/s12264-013-1382-3.
- Kotajima-Murakami H., Kobayashi T., Kashii H., Sato A., Hagino Y., Tanaka M., Nishito Y., Takamatsu Y., Uchino S., Ikeda K. Effects of rapamycin on social interaction deficits and gene expression in mice exposed to valproic acid in utero. Mol Brain 2019; 12(1): 3, https://doi.org/10.1186/s13041-018-0423-2.
- Winden K.D., Ebrahimi-Fakhari D., Sahin M. Abnormal mTOR activation in autism. Annu Rev Neurosci 2018; 41: 1–23, https://doi.org/10.1146/annurev-neuro-080317-061747.
- Dave U.P. Genetics of autism spectrum disorder & BDNF gene. Mol Cytogenet 2013; 7: O3.
- Armeanu M., Mokkonen M., Crespi B. Meta-analysis of BDNF levels in autism. Cell Mol Neurobiol 2017; 37(5): 949–954, https://doi.org/10.1007/s10571-016-0415-7.










