Spectrum of PRSS1, SPINK1, CTRC, CFTR, and CPA1 Gene Variants in Chronic Pancreatitis Patients in Russia
The aim of the study was to define the spectrum of genetic risk factors of chronic pancreatitis (CP) development in patients living in the European part of the Russian Federation.
Materials and Methods. The study group included 105 patients with CP, with the age of the disease onset under 40 years old (the average age of onset was 26.9 years). The control group consisted of 76 persons without clinical signs of pancreatitis. The diagnosis of chronic pancreatitis in patients was made on the basis of clinical manifestations and the results of laboratory and instrumental investigations. Genetic examination of patients was conducted using the next-generation sequencing (NGS) technology and included targeted sequencing of all exons and exon-intron boundaries of the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes. The genotyping of the rs61734659 locus of the PRSS2 gene was also conducted.
Results. Genetic risk factors of the CP development were found in 61% of patients. Pathogenic and likely-pathogenic variants associated with the risk of CP development were identified in the following genes: CTRC (37.1% of patients), CFTR (18.1%), SPINK1 (8.6%), PRSS1 (8.6%), and CPA1 (6.7%). The frequent gene variants in Russian patients with CP were as follows: CTRC gene — c.180C>T (rs497078), c.760C>T (rs121909293), c.738_761del24 (rs746224507); cumulative odds ratio (OR) for all risk alleles was 1.848 (95% CI: 1.054–3.243); CFTR gene — c.3485G>T (rs1800120), c.1521_1523delCTT (p.Phe508del, rs113993960), and c.650A>G (rs121909046); OR=2.432 (95% CI: 1.066–5.553). In the SPINK1, PRSS1, and CPA1 genes, pathogenic variants were found only in the group of patients with CP. The frequent variants of the SPINK1 gene include c.101A>G (p.Asn34Ser, rs17107315) and c.194+2T>C (rs148954387); of the PRSS1 gene — c.86A>T (p.Asn29Ile, rs111033566); of the CPA1 gene — c.586-30C>T (rs782335525) and c.696+23_696+24delGG. The OR for the CP development for the c.180TT genotype (rs497078) CTRC according to the recessive model (TT vs. CT+CC) was 7.05 (95% CI: 0.86–263, p=0.011). In the CTRC gene, the variant c.493+49G>C (rs6679763) appeared to be benign, the c.493+51C>A (rs10803384) variant was frequently detected among both the diseased and healthy persons and did not demonstrate a protective effect. The protective factor c.571G>A (p.Gly191Arg, rs61734659) of the PRSS2 gene was detected only in the group of healthy individuals and confirmed its protective role. 12.4% of the patients with CP had risk factors in 2 or 3 genes.
Conclusion. Sequencing of the coding regions of the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes allowed to identify genetic risk factors of the CP development in 61% of cases. Determining the genetic cause of CP helps to predict the disease course, perform preventive measures in the proband’s relatives, and facilitate a personalized treatment of the patient in future.
Introduction
Chronic pancreatitis (CP) is characterized by a chronic inflammatory process in the pancreatic tissue, which, with the disease progression, is replaced by fibrous tissue. The incidence of CP in adults is 1 per 2632 persons, in children — 1 per 7692 [1, 2]. However, due to various reasons, there may be a certain part of unrecognized cases of this disease.
In general, risk factors contributing to the CP development are well known. They include smoking, excessive alcohol drinking, cholelithiasis, exposure to stress factors, hypertriglyceridemia, hypercalcemia, injuries and malformations of the pancreas, taking of certain medications, etc. [3]. However, in clinical practice, there are often situations when it is not possible to specify the exact cause of the disease. In such cases, “idiopathic pancreatitis” is diagnosed. For these patients, conduction of a genetic examination is of utmost importance, as it is possible to identify a hereditary form of CP in approximately 20% of cases [2, 4].
According to the latest version of the classification of the main CP causes (TIGAR-O Version 2), genetic factors play a significant role in the disease pathogenesis [5].
Products of the following genes: PRSS1 (cationic trypsinogen type 1), SPINK1 (secretory pancreatic inhibitor of trypsin type 1), CTRC (chymotrypsin C), CFTR (cystic fibrosis transmembrane conductance regulator), and CPA1 (carboxypeptidase A1) are essential for the pancreas functioning [4].
The aim of the study was to determine the structure of chronic pancreatitis genetic causes in patients living in the European part of Russia by sequencing the entire coding sequence of the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes.
Materials and Methods
The study group included 105 patients with CP with the age of disease onset under 40 years old (the average age at the time of blood sampling was 35.1 years; the average age of CP manifestation was 26.9 years). Of the patients, 65 were males (average age at the time of blood sampling was 36.3 years; average age of CP onset was 28.7 years) and 40 were females (average age at the time of blood sampling was 33.2 years; average age of CP onset was 23.9 years). The control group included 76 persons of the corresponding gender and age without pancreatitis (average age was 31.6 years).
All study participants provided their voluntary informed consent to participate in the study. The work was approved by the local ethics committee of the Loginov Moscow Clinical Scientific Center of Moscow Healthcare Department (Russia) (protocol No.8 of 2015) and complies with the requirements of the Declaration of Helsinki (2013).
The diagnosis of chronic pancreatitis was made on the basis of clinical manifestations and the results of laboratory and instrument investigations. The genetic study was conducted using the next-generation sequencing (NGS) technology, which included targeted sequencing of all exons and exon-intron bounderies of the PRSS1 (5 exons), SPINK1 (4 exons), CTRC (8 exons), CFTR (27 exons), and CPA1 (10 exons) genes. Moreover, in this study we decided to investigate the role of the protective factor rs61734659 of the PRSS2 gene in relation to the CP development in Russian patients, that is why this gene region was included in the developed targeted genetic panel.
For genetic analysis, DNA was isolated from venous whole blood samples of the study participants using the DNeasy Blood & Tissue Kit (QIAGEN, Germany) on an automated system for the DNA, RNA, and proteins isolation QIAcube (QIAGEN, Germany) according to the manufacturer’s standard protocol.
Sample preparation for sequencing included a PCR test with a primer panel of 67 primer pairs (with a total coverage of 22,693 nucleotides), which was developed specifically for this study with the Ion AmpliSeq Designer software (Thermo Fisher Scientific, USA). The nucleotide sequences of the analyzed genes were received from the NCBI database (https://www.ncbi.nlm.nih.gov/). The targeted amplification was conducted using reagents for PCR tests (AmpliSense; Central Research Institute of Epidemiology of the Russian Federal Service for Surveillance on Consumer Rights Protection and Human Wellbeing, Russia) on a QuantStudio 5 Real-Time PCR Systems machine (Thermo Fisher Scientific, USA).
The NGS libraries were prepared using T4 Polynucleotide Kinase (New England Biolabs, USA) and T4 DNA Ligase (New England Biolabs, USA) reagents according to the manufacturer’s instructions with minor modifications.
At all stages, purification of the PCR product and NGS libraries from components of the reaction mixtures was conducted using carboxylated magnetic particles Sera-Mag SpeedBeads (Sigma-Aldrich, USA). The DNA concentration of the isolated samples, the PCR product, and the final libraries was measured fluorometrically using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, USA) on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, USA). The quality of the final libraries was assessed on a chip by capillary electrophoresis on Agilent 2100 Bioanalyzer (Agilent Technologies, USA) using an Agilent High Sensitivity DNA Kit (Agilent Technologies, USA).
Sequencing on the Ion S5 platform (Thermo Fisher Scientific, USA) was conducted using an Ion 520 & Ion 530 Kit-Chef (Thermo Fisher Scientific, USA) and a semiconductor chip Ion 530™ Chip Kit (Thermo Fisher Scientific, USA).
Bioinformatic processing of the NGS sequencing data included deletion of low-quality reads using the PRINSEQ-lite software [6]; mapping to the reference human genome (GRCh38.p7, PRJNA31257) with Burrows–Wheeler Aligner (BWA-MEM, v_0.7.13) [6]; search for nucleotide sequence variants using the Genome Analysis Toolkit (GATK version 4.0.11.0) software package [7]. SAMtools v_1.3.1 [8] and Picard toolkit v_2.18.17 were used to handle sam/bam files; the VEP software [9] using the 94_GRCh38 cache was applied for primary annotation of variants.
Validation of nucleotide sequence variants detected by automatic analysis was conducted manually using the Tablet graphical user interface for viewing assemblies and alignments [10].
Clinical interpretation of the identified variants was conducted using the following electronic resources and databases: dbSNP (https://www.ncbi.nlm.nih.gov/snp/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), Cystic Fibrosis Mutation Database (http://www.genet.sickkids.on.ca), CFTR2 (https://cftr2.org), CFTR-France (https://cftr.iurc.montp.inserm.fr), OMIM (https://www.omim.org/), pancreasgenetics.org.
Statistical analysis was conducted using the standard R software package [11]. To specify the association between polymorphisms of the analyzed genes and the CP development, the odds ratio (OR) and the relative risk (RR) were calculated using the standard formulas with the calculation of a 95% confidence interval (CI). To determine the significance level of value differences, Student’s t-test was applied with the calculation of χ2 and the p-value of significance. Differences between the samples were considered statistically significant at p≤0.05. The standard deviation of the percentage was calculated by the formula:
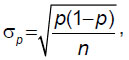
where p is the percentage, σp is the standard deviation of the percentage, and n is the number of assessments.
Results
Variants of the CTRC gene. Most often, genetic risk factors of the disease development were identified in the CTRC gene. In the majority of cases, patients were heterozygous (n=22) or homozygous (n=8) carriers of the so-called risk diplotype c.180C>T (rs497078)/c.493+52G>A (rs545634), which was found in 30 patients. In three more patients with CP, the c.180C>T (rs497078) variant was identified without combination with c.493+52G>A (rs545634): in two cases in the heterozygous and in one case in the homozygous state. Thus, 33 persons out of 105 (31.4%) had the c.180C>T (rs497078) variant of the CTRC gene. The second frequent finding in the CTRC gene in patients with CP was a gene variant of unknown clinical significance — c.493+49G>C (rs6679763), which was identified in five persons in the heterozygous state (4.8%). Four other patients (3.8%) had pathogenic variant c.760C>T (p.Arg254Trp, rs121909293) in the heterozygous state, and two patients (1.9%) had pathogenic variant c.738_761del24 (p.Lys247_Arg254del, rs746224507) in heterozygous state as well. One patient had a gene variant of unknown clinical significance — c.-59C>T (rs183658182) in the heterozygous form (Table 1).

|
Table 1. Frequency of alleles and genotypes of polymorphic loci of the CTRC gene in patients with chronic pancreatitis and healthy individuals |
In the control group, 21 individuals (27.6%) had changes in the CTRC gene in the heterozygous state. Of them, 16 persons were heterozygous carriers of the c.180C>T (rs497078) gene variant in the form of the diplotype c.180C>T (rs497078)/c.493+52G>A (rs545634) variant, four persons were heterozygous carriers of the c.180C>T variant (rs497078) without c.493+52G>A (rs545634) variant, one person had the c.760C>T (p.Arg254Trp, rs121909293) gene variant (see Table 1).
Comparison of the frequency of c.180T allele (rs497078) in the group of patients with CP (20%) and the control group (13.2%) showed that differences were statistically insignificant (χ2=2.908; p=0.089). However, homozygosity for the c.180C>T (rs497078)/c.493+52G>A (rs545634) diplotype was observed only in patients with CP (see Table 1). This indicates a significant effect of the homozygous genotype for the c.180C>T variant (rs497078) of the CTRC gene on the development of inflammatory changes in the pancreas. The OR for the c.180TT genotype according to the recessive model (TT vs. CT+CC) was 7.05 (95% CI: 0.86–263, p=0.011).
It is of interest to note that the c.493+49G>C variant (rs6679763) was identified in a heterozygous form in 8 out of 76 healthy persons (10.5%). When comparing the frequencies of different genotypes and alleles for the rs6679763 polymorphic locus, no statistically significant differences were detected between the group of patients and the control group (p=0.139 and p=0.146, respectively). This fact leads to a conclusion that this variant has no pathogenic effect on the risk of the CP development.
The variant of unknown clinical significance c.-59C>T (rs183658182) in the CTRC gene was identified in a heterozygous form in one person from the control group, which can be a sign of its low significance for the CP development. The c.760C>T variant (p.Arg254Trp, rs121909293) was found in one person from the control group (a 23-year-old woman without manifestations of CP at the time of blood sampling). The c.738_761del24 variant (p.Lys247_Arg254del, rs746224507/rs515726210) was not detected in individuals from the control group.
In addition to the above-mentioned findings, both in the group of patients with CP and in the control group we found the CTRC gene frequent polymorphism c.493+51C>A (rs10803384), which, according to the information from the pancreasgenetics.org database, has a protective role in the disease development. In this study, no significant differences in the frequency of genotypes and alleles for this CTRC polymorphic locus were identified between the two groups (see Table 1).
To summarize, pathogenic variants c.760C>T (p.Arg254Trp, rs121909293) and c.738_761del24 (p.Lys247_Arg254del, rs746224507) and genetic risk factors c.180C>T (rs497078)/c.493+52G>A (rs545634) of the CTRC gene were detected in 39 out of 105 examined patients with CP (37.1%).
The most frequent and significant variants of the CTRC gene identified in patients with CP were as follows: the c.180C>T (rs497078)/c.493+52G>A (rs545634) diplotype and the c.180C>T (rs497078) variant — 84.6% of all findings in this gene; c.760C>T (p.Arg254Trp, rs121909293) — 10.3% of all findings in this gene; c.738_761del24 (p.Lys247_Arg254del, rs746224507) — 5.1% of all findings in this gene.
The frequency of detection of any of the above-mentioned risk alleles of the CTRC gene in the group of patients with CP was 22.9% (48 alleles out of 210), whereas the same parameter for the control group was 13.8% (21 alleles out of 152) (χ2=4.672; p=0.031). The OR for CP development in case of the risk allele of the CTRC gene was 1.848 (95% CI: 1.054–3.243; standard error of the mean — 0.287).
Variants of the CFTR gene. 19 out of 105 patients with CP (18.1%) had pathogenic or likely-pathogenic variants of the CFTR gene in different combinations: c.2620-6T>C, c.224G>A (p.Arg75Gln), c.451C>A (p.Gln151Lys), c.650A>G (p.Glu217Gly), c.1079C>T (p.Thr360Ile), c.2012delT (p.Leu671Ter), c.1516A>G (p.Ile506Val), c.1521_1523delCTT (p.Phe508del), c.1399C>T (p.Leu467Phe), c.1584G>A (p.Glu528=), c.3485G>T (p.Arg1162Leu), c.2991G>C (p.Leu997Phe), c.1969A>G (p.Arg657Gly), c.2619+86delT. Three pathogenic variants, c.3485G>T (p.Arg1162Leu), c.1521_1523delCTT (p.Phe508del), and c.650A>G (p.Glu217Gly), were repetitive and were identified in seven patients (6.7%). One of the CFTR gene variants (c.2619+86delT) was detected for the first time in the study in five patients (in all cases, in the homozygous form). More detailed information about the patients and the variants of the CFTR gene identified in them can be found in our earlier publication [12].
In the control group, pathogenic or likely-pathogenic variants of the CFTR gene were also identified. For instance, 8 out of 76 (10.5%) individuals without symptoms of pancreatitis had variants c.650A>G (p.Glu217Gly), c.3854C>T (p.Ala1285Val), c.443T>C (p.Ile148Thr), c.1584G>A (p.Glu528=), c.224G>A (p.Arg75Gln), c.3532_3535dup (p.Thr1179IlefsTer17) in the heterozygous state.
The frequency of detection of the pathologically altered allele of the CFTR gene in the group of patients with CP was 11.9% (25 alleles out of 210), whereas the same parameter for the control group was 5.3% (8 alleles out of 152) (χ2=4.695; p=0.031). The OR of the CP development in case of the presence of the pathological allele of the CFTR gene in the genotype was 2.432 (95% CI: 1.066–5.553; standard error of the mean — 0.421).
Variants of the SPINK1 gene. Pathogenic and likely-pathogenic variants of the SPINK1 gene were identified only in the group of patients with CP (all variants were found in the heterozygous state). Eight patients (7.6%) were carriers of the known pathogenic variants of the SPINK1 gene: 6 patients had pathogenic variant c.101A>G (p.Asn34Ser, rs17107315) combined with the risk factor c.56-37T>C (rs17107318), 2 patients had variant c.194+2T>C (rs148954387) (Table 2).

|
Table 2. Characteristics of the SPINK1 gene variants in Russian patients with chronic pancreatitis |
Moreover, a variant of unknown clinical significance c.126A>G (p.Ile42Met, rs370266754) was found in one case, and the c.88-23A>T (rs199929811) variant was identified in two patients. The latter variant, according to the pancreasgenetics.org database, is benign and does not increase the risk of CP development. In our study, this variant (rs199929811) was not identified in the control group, which leads to the need to revise the data on the clinical interpretation of this variant.
Therefore, without the benign variant rs199929811 of the SPINK1 gene, 9 out of 105 patients with CP (8.6%) were carriers of pathogenic variants. The most frequent variants include: c.101A>G (p.Asn34Ser, rs17107315) combined with the risk factor c.56-37T>C (rs17107318) — 66.7% of all findings in this gene and c.194+2T>C (rs148954387) — 22.2% of all findings in this gene.
Variants of the PRSS1 gene. Similar to the SPINK1 gene, pathogenic and likely-pathogenic variants of the PRSS1 gene were identified only in the group of patients with CP (all of them are in the heterozygous state).
5 out of 105 patients with CP (4.8%) had known pathogenic variants of the PRSS1 gene: 3 patients were carriers of the c.86A>T (p.Asn29Ile, rs111033566) variant, 1 patient had the c.365G>A (p.Arg122His, rs111033565) variant, and 1 more patient had variant c.68A>G (p.Lys23Arg, rs111033567) (Table 3).

|
Table 3. Characteristics of the PRSS1 gene variants in Russian patients with chronic pancreatitis |
Moreover, there were additional findings during the sequencing of the PRSS1 gene in the group of patients with CP. Variant c.455-93T>C (rs569672741) was identified in the heterozygous form in two patients. According to the information from the dbSNP database, the frequency of the c.455-93C allele in the European population is 0.1%. In the ClinVar database and in the specialized database pancreasgenetics.org, this variant is not annotated. It should be noted that this PRSS1 gene substitution was not found in this study in the control group.
A similar situation was observed for the PRSS1 intron variant c.200+32C>T (rs182426777), which was identified in the heterozygous form in two patients with CP. This variant is not described in the mentioned databases and occurs in the European population with a frequency of approximately 0.3%.
Thus, c.86A>T (p.Asn29Ile, rs111033566) was found in three patients and can be called a major pathogenic variant of the PRSS1 gene in Russian patients with CP.
The cumulative frequency of risk alleles of the PRSS1 gene in the group of patients with CP was 4.3% (9 alleles); in the control group, it was null (χ2=6.680; p=0.01).
Variants of the CPA1 gene. Sequencing of the entire coding sequence of the CPA1 gene revealed several variants that require additional studying.
Two out of 105 patients with CP (1.9%) were heterozygous carriers of the c.586-30C>T variant (rs782335525), which is not annotated in the ClinVar and pancreasgenetics.org databases and was not found in the control samples. This variant occurs in the European population with a frequency of 0.015%.
One patient had the CPA1 c.-90C>T variant (rs377553060) in the heterozygous state, which was also not described in the mentioned databases and was not found in the control group. This variant occurs in the European population with a frequency of 0.03%.
Three unrelated patients with CP (2.8%) had the c.696+23_696+24delGG variant (chr7 130383816 TGG>TG,T) in the homozygous form. Previously this variant was not described in the literature, it is not annotated in genomic databases, and its frequency is not established. It was not registered in the control samples in this study. Segregation genetic analysis in the families of patients was not conducted.
One patient with CP was a heterozygous carrier of the CPA1 c.983A>G (p.Glu328Gly) missense variant, which, similar to the previous variant, was identified for the first time in this cohort of patients with CP.
In two patients with CP (1.9%), the c.509_510insGG (p.Ala171GlufsTer121) (chr7 130383416 C>CGG) variant was identified in the heterozygous form. This variant of the CPA1 gene is not annotated in the global genomic databases. It should be noted that pathogenic variants of the missense type with uncertain clinical significance were previously described in this region of the CPA1 gene. However, in this study, the c.509_510insGG variant was also identified in a heterozygous state in 5 out of 76 healthy persons of the control group (6.6%), which indicates that there is no association between its presence and an increased risk of pancreatitis development (Table 4).

|
Table 4. Characteristics of the CPA1 gene variants in Russian patients with chronic pancreatitis |
Thus, the total frequency of potentially risk alleles of the CPA1 gene in the group of patients with CP was 4.8% (10 alleles); in the control group, it was 0% (χ2=7.444; p=0.007).
Protective factor of the PRSS2 gene. The protective factor c.571G>A (p.Gly191Arg, rs61734659) of the PRSS2 gene was identified only in the group of healthy individuals, seven of them were heterozygous carriers of this polymorphism. Thus, the frequency of the c.571A allele in the group of patients with CP was 0%, whereas in the control group it was 4.6% (χ2=9.862; p=0.002). That means that the protective effect of the c.571A allele at the rs61734659 polymorphic locus was confirmed.
Summary data on identified genetic risk factors. 26.5% of patients with CP had a family history of this disease.
As a result of the genetic testing, in 64 out of 105 patients (61%) with the age of CP onset under 40 years old, significant molesular-genetic findings were identified in different combinations in the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes.
Summary data on the results of sequencing with the use of the developed targeted genetic panel is shown in Table 5.

|
Table 5. Frequency of identification of genetic risk factors of chronic pancreatitis development with the use of the used targeted genetic panel |
Most frequently, patients with CP had CTRC gene variants (37.1% of patients). Less frequently we saw pathogenic and likely-pathogenic variants of the CFTR (18.1%), SPINK1 (8.6%), PRSS1 (8.6%), and CPA1 (6.7%) genes.
The contribution of PRSS1, SPINK1, CTRC, CFTR, and CPA1 gene variants to the development of CP in a cohort of Russian patients is demonstrated in Table 6, which shows that the OR for the CP development for the CTRC and CFTR gene variants is 1.848 and 2.432, respectively. The maximum risks of developing chronic inflammation in the pancreas are identified for carriers of pathogenic variants in the PRSS1, SPINK1, and CPA1 genes, which is consistent with the currently accepted autosomal dominant theory of inheritance of PRSS1- and SPINK1-associated CP (OMIM #167800).

|
Table 6. Association of the risk alleles of PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes with chronic pancreatitis development in the Russian cohort of patients |
Some patients with CP had a combination of pathogenic variants and risk factors in several genes at the same time. For instance, 11 patients were carriers of two genetic risk factors, two patients had three genetic risk factors (Table 7).

|
Table 7. Combination of genetic risk factors in patients with chronic pancreatitis |
Discussion
The present study, which included 105 patients with CP, was characterized by a specific predominance of males, which is consistent with the data from the literature, according to which the ratio of men and women among patients with CP is 1.05 [13].
Based on the assumption that hereditary forms of CP manifest at an earlier age, the group of the study participants included patients with an age of the disease onset under 40 years old. There are publications in the worldwide literature where the authors used the same principle to form the group of the patients for the further investigation [14–16]. For instance, Zou et al. [17] showed that the average age of pancreatitis onset in patients with molecular findings in the SPINK1, PRSS1, CTRC, and CFTR genes was 29.70±14.84 years, whereas in patients having no mutations in these genes the first clinical signs of the disease appeared at the age of 43.01±15.97 years [17].
The implementation of this approach allowed to identify genetic predictors of the CP development in the majority (61%) of patients, which is generally consistent with the literature. In particular, when sequencing the SPINK1, PRSS1, CTRC, and CFTR genes in a large group of patients with CP, Zou et al. [17] identified pathogenic variants in 50.42% of cases. At that, mutations of the mentioned genes were detected by the authors only in 5.94% of cases (OR=16.12; p<0.001) in the control group [17].
The content of the genetic panel developed in this study is influenced by the results of original studies and data from genomic databases, according to which the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes play a significant role in molecular pathogenesis of the disease [18]. The PRSS1 gene is responsible for production of cationic trypsinogen type 1, the SPINK1 gene is a secretory pancreatic inhibitor of trypsin type 1, the CTRC gene is chymotrypsin C, the CFTR gene is a cystic fibrosis transmembrane conductance regulator, the CPA1 gene is associated with abnormal clotting of the carboxypeptidase A1 protein and endoplasmic stress of reticulum in pancreatic cells [4]. It should be noted that hereditary forms of CP associated with mutations of the PRSS1 (MIM# 276000) and SPINK1 (MIM# 167790) genes are characterized by an autosomal dominant type of inheritance.
The results of the analysis of the entire coding sequence of these genes using NGS to identify major pathogenic variants and genetic risk factors specific to the Russian population are of interest. Until now, patients with CP in the Russian Federation have not been genotyped so comprehensively. Currently, in most cases, the genetic diagnostics of CP in Russia is limited to finding several frequent mutations in the PRSS1 and SPINK1 genes. In certain cases, patients undergo sequencing of the coding regions of the PRSS1 and SPINK1 genes, which, according to the data of this study, is insufficient and may omit the CP genetic risk factors in patients in the majority of cases.
Moreover, the results of this study allowed to form a list of genetic markers which can be used as screening for the most common genetic risk factors of CP development in Russian patients.
The most frequent molecular finding among patients with CP were CTRC gene variants, which were identified in 37.1% of patients and in this population might be considered as risk factors (see Table 5). The OR of pathology development in case of genetic risk factors of this gene was 1.848 (p=0.031). At the same time, different variants of this gene contribute to the risk of the CP development in different manner.
The c.180C>T (rs497078)/c.493+52G>A (rs545634) diplotype of the CTRC gene is widely discussed in the literature and is a known genetic risk factor of the CP development. For instance, LaRusch et al. [19] demonstrated an association of the c.180T allele (rs497078) with the risk of CP development in the European population (OR for the CT genotype was 1.36; for the TT genotype — 3.98). This study identified an association of the c.180C>T (rs497078) polymorphism with the increased risk of CP development only in case of homozygosity for this variant (OR=7.05; p=0.011). The lack of a statistically significant association of rs497078 in heterozygous state with an increased probability of pathology development in this study might be due to the characteristics of the cohort, and particularly to the early CP onset in patients, as a later disease onset is typical for individuals with pathogenic variants of this gene.
Other changes in the CTRC gene in patients with CP have a greater impact on the risk of the disease development. For instance, the pathogenic variant c.760C>T (p.Arg254Trp, rs121909293) was found in the heterozygous state in four (3.8%) patients with CP and only in one (1.3%) person from the control group. However, this control sample was obtained from a 23-year-old woman who, due to her age, might not yet have CP onset. These findings correlate with the data of foreign researchers, in particular, with the results of study by Rosendahl et al. [20], in which the frequency of this missense variant in a cohort of German patients with idiopathic and hereditary CP was 2.1%, whereas in the control group it was identified only in 0.6% of cases.
A similar situation was observed for the pathogenic variant c.738_761del24 (p.Lys247_Arg254del, rs746224507/rs515726210), which was identified in the study only in the group of patients with CP.
Therefore, a relatively rare but recurring variants c.760C>T (p.Arg254Trp, rs121909293) and c.738_761del24 (p.Lys247_Arg254del, rs515726210) have the maximum impact on the risk of CP development in the study cohort.
Moreover, we demonstrated that the c.493+49G>C (rs6679763) variant of the CTRC gene, which is described as a variant of unknown significance in the specialized database on genetic findings in CP (pancreasgenetics.org), did not increase the risk of the disease development and therefore was a benign variant.
A frequent variant of the CTRC gene, c.493+51C>A (rs10803384), described in the pancreasgenetics.org database as protective factor regarding the CP development, did not show a protective effect in the current study.
Another variant of the CTRC gene of unknown clinical significance, c.-59C>T (rs183658182), requires further investigation, and this study’s results cannot attribute it to the risk factors.
Findings in the CFTR gene in the group of patients with CP were the second in terms of prevalence. 18.1% of patients were carriers of various variants of this gene. Among all variants, the following are recurring in Russian patients with CP: c.3485G>T (p.Arg1162Leu), c.1521_1523delCTT (p.Phe508del), and c.650A>G (p.Glu217Gly). One of the variants of the CFTR gene (c.2619+86delT), first discovered in this study in five patients with CP in the homozygous form, requires further investigation in terms of both prevalence and functional significance. Detailed information on CFTR gene variants and patients with CFTR-related CP can be found in the article published by Litvinova et al. [12]. The cumulative OR of the CP development with a pathological CFTR allele was 2.432, which in general is consistent with the study by Zou et al. [17], where the same parameter was 3.71.
Pathogenic changes in the SPINK1 and PRSS1 genes were identified in 8.6% of patients with CP. The present data differ from the level of detection of mutations in these two genes shown by Masamune et al. [21], where 41% of patients were carriers of pathogenic variants of the PRSS1 gene and 36% of patients had mutations in the SPINK1 gene. Potentially, this difference is due to the fact that the present study did not focus on selection of patients with a family history of pancreatitis, whereas the mentioned study conducted in Japan (231 samples) included patients with a family history of CP.
Pathogenic variants of the SPINK1 and PRSS1 genes have the maximum impact on the risk of CP development in patients from different countries, as the products of these genes are key participants in the trypsin-dependent pathogenetic pathway of the pancreatitis development. Very often, patients with mutations in the SPINK1 and PRSS1 genes have a family history of CP in several generations, moreover, the age of the disease onset is usually much earlier compared to CFTR- or CTCR-associated CP [17]. Comparison of the current study results with similar studies conducted by foreign researchers revealed a rather large similarity in the spectrum of genetic findings in the SPINK1 and PRSS1 genes in the Russian and European patient populations. The most common pathogenic variants of the SPINK1 gene in the Russian cohort include the missense variant c.101A>G (p.Asn34Ser, rs17107315) and the splicing site mutation c.194+2T>C (rs148954387); the recurrent pathogenic variant of PRSS1 is c.86A>T (p.Asn29Ile, rs111033566), which, according to foreign researchers, occur in 5–21% of patients with this disease [4].
Specific variants of the SPINK1 and PRSS1 genes were also highlighted as requiring further study (SPINK1: rs370266754, rs199929811; PRSS1: rs569672741, rs182426777). For instance, the c.88-23A>T (rs199929811) variant of the SPINK1 gene is annotated in the databases as a benign variant, but the results of the current study require revising clinical interpretation of this variant, as it was identified only in patients and was not found in anyone from the control group. A similar situation was seen for the rs569672741 and rs182426777 variants of the PRSS1 gene.
Currently, the CPA1 gene is the least studied among CP-predisposing genes. However, the results of the functional analysis of various changes in CPA1 in the cell cultures and model animals indicate a significant contribution of some variants of this gene to the pathogenesis of the disease [22]. According to [4], CPA1 gene variants in patients lead to an earlier onset of the disease. According to the results of our study, the following variants of the CPA1 gene must be paid the greatest attention: c.586-30C>T (rs782335525), c.-90C>T (rs377553060), c.983A>G (p.Glu328Gly), and c.696+23_696+24delGG (chr7 130383816 TGG>TG,T). We are planning to conduct further study of these variants in the Russian cohort of patients with CP.
Therefore, the major contribution of the genetic causes of CP in Russian patients was observed (in descending order) for the CTRC gene variants (identified in 37.1% of cases), CFTR (18.1%), SPINK1 (8.6%), PRSS1 (8.6%), and CPA1 (6.7%). This data is quite consistent with the results of the study in the European cohort of patients with CP [23]. At the same time, a large study in China revealed specific details that demonstrated the maximum impact of SPINK1 (identified in 61.5%) and PRSS1 (13.5%) gene variants on the risk of idiopathic pancreatitis development. The CTRC and CFTR genes variants were found in only 1.5 and 4.9% of cases, respectively [17]. This may prove the fact that the genetic diagnostics of CP should be performed taking into account the population characteristics. It is essential to pay attention to such characteristics during the development of genetic tests for genetic analysis of CP.
We could not find a genetic causative factors in 39% of CP patients, which is generally consistent with the data of foreign researchers [24]. Patients having no genetic risk factors identified during genetic testing might have additional genetic factors associated both with the polygenic nature of the pathology and with monogenic form of the disease, the clinical manifestations of which can include inflammation of the pancreatic tissue. For instance, it is known that some monogenic metabolic disorders (methylmalonic aciduria, impaired fatty acid oxidation, etc.) are characterized by clinical signs of CP in childhood and adolescence [25]. Meanwhile, identification of the exact genetic cause of the disease is of utmost importance.
Currently, the genetic characteristics of patients with CP allow to make a conclusion about the peculiarities of the course of the disease, its prognosis, and the probability of specific complications development, which primarily include pancreatic exocrine insufficiency, diabetes mellitus, and pancreatic cancer [26]. Taking into account the fact of active introduction of gene therapeutic technics and targeted therapy aimed to influence on the specific points of the molecular pathogenesis of the disease into clinical practice, it is obvious that the future will provide new opportunities for treatment of patients with CP based on their genetic profile. For instance, it was demonstrated that CFTR protein potentiators can reduce the number of pancreatitis attacks in patients with CFTR-associated pancreatitis [27].
Conclusion
The developed genetic panel (PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes) for identification of genetic risk factors of the chronic pancreatitis development and the usage of the next-generation sequencing technology allowed to specify genetic predictors of the disease development in 61% of patients. Pathogenic and potentially pathogenic variants associated with the risk of this disease development were identified in the following genes: CTRC (37.1% of patients), CFTR (18.1%), SPINK1 (8.6%), PRSS1 (8.6%), and CPA1 (6.7%). Recurrent variants of the PRSS1, SPINK1, CTRC, CFTR, and CPA1 genes, contributing to the genetic predisposition to chronic pancreatitis development were identified in a cohort of Russian patients. It is assumed that the study results will be applied in genetic diagnostics and interpretation of the results of molecular genetic tests in chronic pancreatitis patients in the Russian Federation, as well as will help to increase the effectiveness of genetic analysis for families with chronic diseases of the gastrointestinal tract and, in particular, with diseases of the pancreas.
Study funding. The study was not funded by any sources.
Conflicts of interest. The authors of the publication claimed that they had no conflicts of interest to report.
References
- Meyer A., Coffey M.J., Oliver M.R., Ooi C.Y. Contrasts and comparisons between childhood and adult onset acute pancreatitis. Pancreatology 2013; 13(4): 429–435, https://doi.org/10.1016/j.pan.2013.06.005.
- Machicado J.D., Yadav D. Epidemiology of recurrent acute and chronic pancreatitis: similarities and differences. Dig Dis Sci 2017; 62(7): 1683–1691, https://doi.org/10.1007/s10620-017-4510-5.
- Beyer G., Habtezion A., Werner J., Lerch M.M., Mayerle J. Chronic pancreatitis. Lancet 2020; 396(10249): 499–512, https://doi.org/10.1016/s0140-6736(20)31318-0.
- Litvinova M.M., Khafizov K.F., Shipulin G.A., Ayginin A.A., Vinokurova L.V., Nikolskaya K.A., Dubtsova E.A., Bordin D.S., Asanov A.Yu. Genetic factors of the development of chronic pancreatitis. Voprosy prakticeskoj pediatrii 2018; 13(3): 29–40, https://doi.org/10.20953/1817-7646-2018-3-29-40.
- Whitcomb D.C.; North American Pancreatitis Study Group. Pancreatitis: TIGAR-O Version 2 Risk/Etiology Checklist with topic reviews, updates, and use primers. Clin Transl Gastroenterol 2019; 10(6): e00027, https://doi.org/10.14309/ctg.0000000000000027.
- Schmieder R., Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27(6): 863–864, https://doi.org/10.1093/bioinformatics/btr026.
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010; 20(9): 1297–1303, https://doi.org/10.1101/gr.107524.110.
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25(16): 2078–2079, https://doi.org/10.1093/bioinformatics/btp352.
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The Ensembl variant effect predictor. Genome Biol 2016; 17(1): 122, https://doi.org/10.1186/s13059-016-0974-4.
- Milne I., Stephen G., Bayer M., Cock P.J., Pritchard L., Cardle L., Shaw P.D., Marshall D. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform 2013; 14(2): 193–202, https://doi.org/10.1093/bib/bbs012.
- R Foundation. The R project for statistical computing. Vienna; 2019. URL: https://www.R-project.org/.
- Litvinova M.M., Khafizov K.F., Speranskaya A.S., Matsvay A.D., Nikolskaya K.A., Vinokurova L.V., Dubtsova E.A., Muhina T.F., Khavkin A.I., Bordin D.S. Spectrum of CFTR gene mutations in patients with chronic pancreatitis in Russia. Voprosy detskoj dietologii 2020; 18(3): 5–18, https://doi.org/10.20953/1727-5784-2020-3-5-18.
- Machicado J.D., Dudekula A., Tang G., Xu H., Wu B.U., Forsmark C.E., Yadav D. Period prevalence of chronic pancreatitis diagnosis from 2001–2013 in the commercially insured population of the United States. Pancreatology 2019; 19(6): 813–818, https://doi.org/10.1016/j.pan.2019.07.003.
- Saito N., Suzuki M., Sakurai Y., Nakano S., Naritaka N., Minowa K., Sai J.K., Shimizu T. Genetic analysis of Japanese children with acute recurrent and chronic pancreatitis. J Pediatr Gastroenterol Nutr 2016; 63(4): 431–436, https://doi.org/10.1097/mpg.0000000000001320.
- Xiao Y., Yuan W., Yu B., Guo Y., Xu X., Wang X., Yu Y., Yu Y., Gong B., Xu C. Targeted gene next-generation sequencing in Chinese children with chronic pancreatitis and acute recurrent pancreatitis. J Pediatr 2017; 191: 158–163.e3, https://doi.org/10.1016/j.jpeds.2017.08.063.
- Párniczky A., Mosztbacher D., Zsoldos F., Tóth A., Lásztity N., Hegyi P.; Hungarian Pancreatic Study Group and the International Association of Pancreatology. Analysis of pediatric pancreatitis (APPLE trial): pre-study protocol of a multinational prospective clinical trial. Digestion 2016; 93(2): 105–110, https://doi.org/10.1159/000441353.
- Zou W.B., Tang X.Y., Zhou D.Z., Qian Y.Y., Hu L.H., Yu F.F., Yu D., Wu H., Deng S.J., Lin J.H., Zhao A.J., Zhao Z.H., Wu H.Y., Zhu J.H., Qian W., Wang L., Xin L., Wang M.J., Wang L.J., Fang X., He L., Masson E., Cooper D.N., Férec C., Li Z.S., Chen J.M., Liao Z. SPINK1, PRSS1, CTRC, and CFTR genotypes influence disease onset and clinical outcomes in chronic pancreatitis. Clin Transl Gastroenterol 2018; 9(11): 204, https://doi.org/10.1038/s41424-018-0069-5.
- Mayerle J., Sendler M., Hegyi E., Beyer G., Lerch M.M., Sahin-Tóth M. Genetics, cell biology, and pathophysiology of pancreatitis. Gastroenterology 2019; 156(7): 1951–1968.e1, https://doi.org/10.1053/j.gastro.2018.11.081.
- LaRusch J., Lozano-Leon A., Stello K., Moore A., Muddana V., O’Connell M., Diergaarde B., Yadav D., Whitcomb D.C. The common chymotrypsinogen C (CTRC) variant G60G (C.180T) increases risk of chronic pancreatitis but not recurrent acute pancreatitis in a North American population. Clin Transl Gastroenterol 2015; 6(1): e68, https://doi.org/10.1038/ctg.2014.13.
- Rosendahl J., Witt H., Szmola R., Bhatia E., Ozsvári B., Landt O., Schulz H.U., Gress T.M., Pfützer R., Löhr M., Kovacs P., Blüher M., Stumvoll M., Choudhuri G., Hegyi P., te Morsche R.H.M., Drenth J.P., Truninger K., Macek M. Jr., Puhl G., Witt U., Schmidt H., Büning C., Ockenga J., Kage A., Groneberg D.A., Nickel R., Berg T., Wiedenmann B., Bödeker H., Keim V., Mössner J., Teich N., Sahin-Tóth M. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet 2008; 40(1): 78–82, https://doi.org/10.1038/ng.2007.44.
- Masamune A., Kikuta K., Hamada S., Nakano E., Kume K., Inui A., Shimizu T., Takeyama Y., Nio M., Shimosegawa T. Nationwide survey of hereditary pancreatitis in Japan. J Gastroenterol 2018; 53(1): 152–160, https://doi.org/10.1007/s00535-017-1388-0.
- Németh B.C., Demcsák A., Geisz A., Sahin-Tóth M. Misfolding-induced chronic pancreatitis in CPA1 N256K mutant mice is unaffected by global deletion of Ddit3/Chop. Sci Rep 2022; 12(1): 6357, https://doi.org/10.1038/s41598-022-09595-x.
- Masson E., Chen J.M., Audrézet M.P., Cooper D.N., Férec C. A conservative assessment of the major genetic causes of idiopathic chronic pancreatitis: data from a comprehensive analysis of PRSS1, SPINK1, CTRC and CFTR genes in 253 young French patients. PLoS One 2013; 8(8): e73522, https://doi.org/10.1371/journal.pone.0073522.
- Masamune A. Genetics of pancreatitis: the 2014 update. Tohoku J Exp Med 2014; 232(2): 69–77, https://doi.org/10.1620/tjem.232.69.
- Hwang W.J., Lim H.H., Kim Y.M., Chang M.Y., Kil H.R., Kim J.Y., Song W.J., Levy H.L., Kim S.Z. Pancreatic involvement in patients with inborn errors of metabolism. Orphanet J Rare Dis 2021; 16(1): 37, https://doi.org/10.1186/s13023-021-01685-9.
- Suzuki M., Minowa K., Nakano S., Isayama H., Shimizu T. Genetic abnormalities in pancreatitis: an update on diagnosis, clinical features, and treatment. Diagnostics (Basel) 2020; 11(1): 31, https://doi.org/10.3390/diagnostics11010031.
- Carrion A., Borowitz D.S., Freedman S.D., Siracusa C.M., Goralski J.L., Hadjiliadis D., Srinivasan S., Stokes D.C. Reduction of recurrence risk of pancreatitis in cystic fibrosis with ivacaftor: case series. J Pediatr Gastroenterol Nutr 2018; 66(3): 451–454, https://doi.org/10.1097/mpg.0000000000001788.










